
Loving Boston (yes, the snow as well). If you are around and work on similar things, or just passionate about science and would like to grab coffee and chat, DM me.
#postdoclife

Loving Boston (yes, the snow as well). If you are around and work on similar things, or just passionate about science and would like to grab coffee and chat, DM me.
#postdoclife
Been a while, finished my PhD last March, moved to Boston in April, and excited about starting as a postdoc in the Buenrostro lab at the Broad.
I will be continuing within hematopoiesis, this time asking how psychosocial stress and resultant endocrine signalling shape HSC fate.
Bonus: you won’t mind Boston humor (it’s a acquired taste). Happy to chat about the role + projects in the lab + the city, speaking as someone with ~8 months of Boston experience.
This could be a great fit if you’re graduating and want ~2 years of research experience before PhD.
Come join us 🙂 The Buenrostro lab at the Broad is looking for a few Research Associates to start ~Summer 2026.
If you’re comfortable with rodents, excited about single-cell tech, and weirdly into hematopoiesis… you’ll fit right in.
www.buenrostrolab.com
Grant writing is such a humbling experience, you read enough to think you can write… then you start writing, just to realise, how reading more might not be a bad idea too, lol..
14.02.2026 23:07 — 👍 0 🔁 0 💬 0 📌 0
Nature research paper: Basal cell of origin resolves neuroendocrine–tuft lineage plasticity in cancer
go.nature.com/3ViRTAP

Now online in Cancer Discovery @aacrjournals.bsky.social: Development and Prospective Validation of a Cell-free DNA-based Model for the Early Detection of Pancreatic Cancer - by Xiuchao Wang, Hongwei Wang, Meng Zhang, Jun Yu, Chuntao Gao, Yunfeng Cui, Jihui Hao, et al. doi.org/10.1158/2159...
24.09.2025 13:43 — 👍 5 🔁 3 💬 0 📌 0
🚨 New preprints from our lab! First, we introduce Cryo-mtscATAC-seq, led by Maren (@ms-maren.bsky.social ), enabling high-throughput clonal tracing from frozen human samples by isolating nuclei with their mitochondria (“CryoCells”). 👉 www.biorxiv.org/content/10.1...
24.09.2025 12:39 — 👍 23 🔁 11 💬 2 📌 1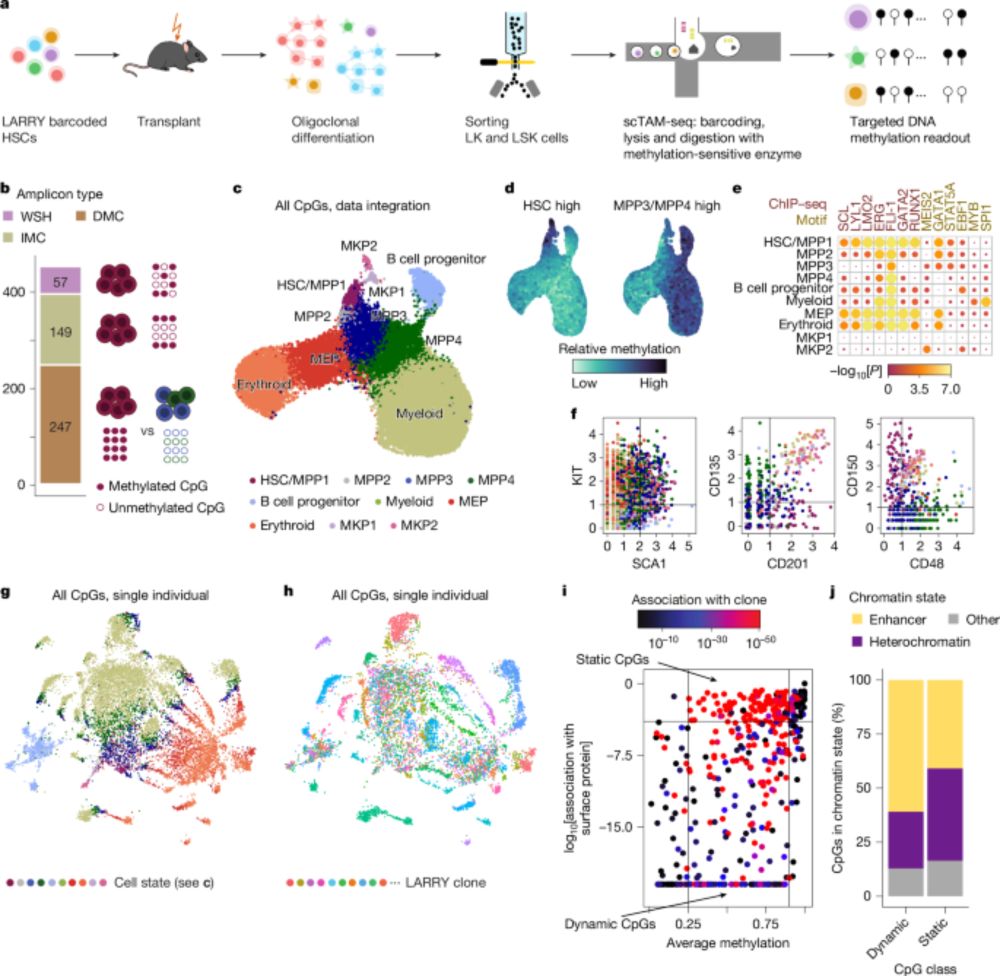
EPI-Clone is a transgene-free lineage tracing method that uses single-cell DNA methylation analysis to track hematopoietic stem cell clones over time #NBThighlight www.nature.com/articles/s41...
23.05.2025 20:05 — 👍 9 🔁 4 💬 0 📌 0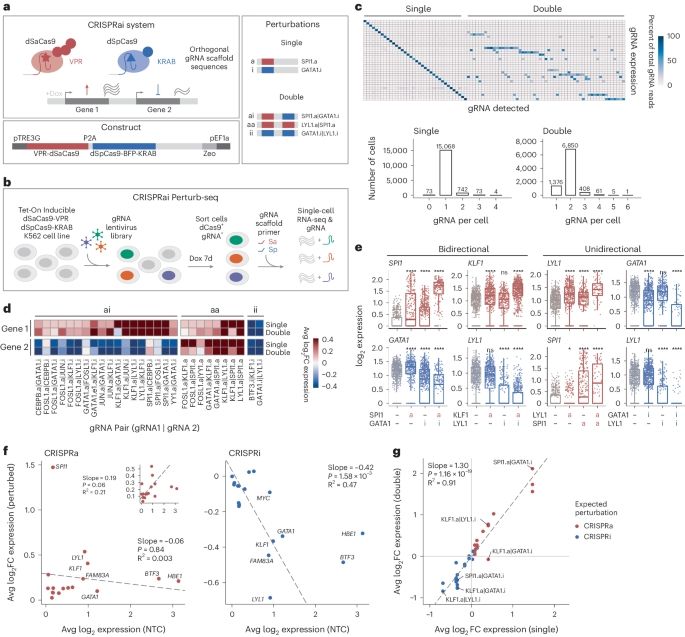
CRISPRai simultaneously activates and represses two genes in single cells go.nature.com/4apFU9q
rdcu.be/efe0E
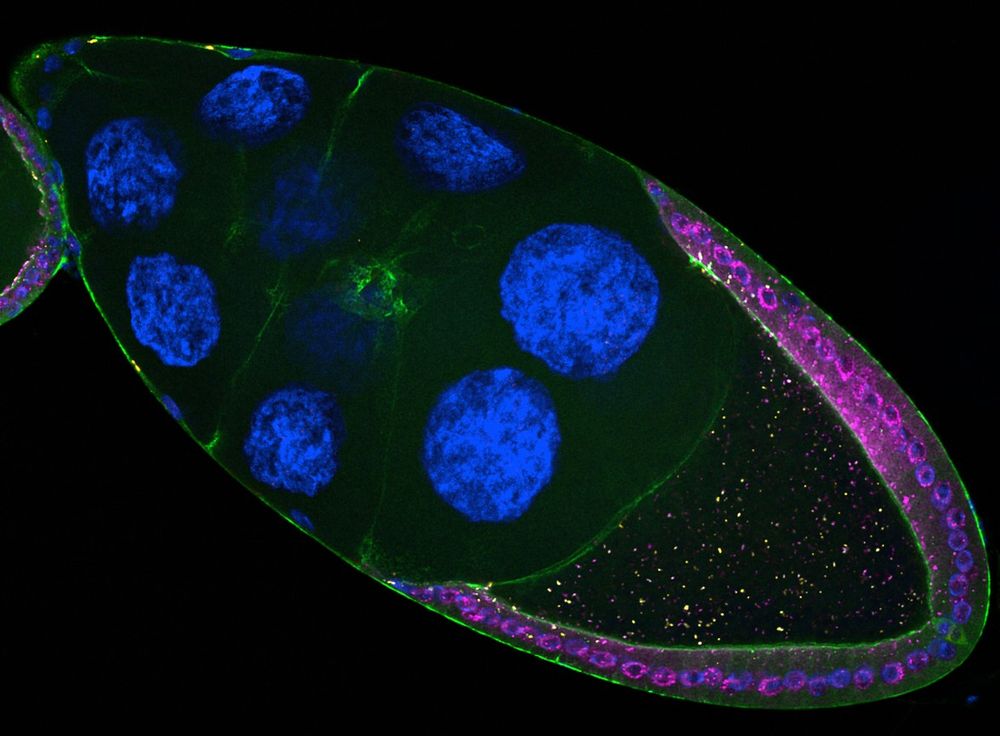
Drosophila follicle showing retrotransposons (pink & yellow) expressed in somatic cells infecting the oocyte
1/ Transposable elements are often called "jumping genes" because they mobilize within genomes. 🧬
But did you know they can also jump 𝘣𝘦𝘵𝘸𝘦𝘦𝘯 cells? 🤯
Our new study reveals how retrotransposons invade the germline directly from somatic cells.
www.biorxiv.org/content/10.1...
A short thread 🧵👇
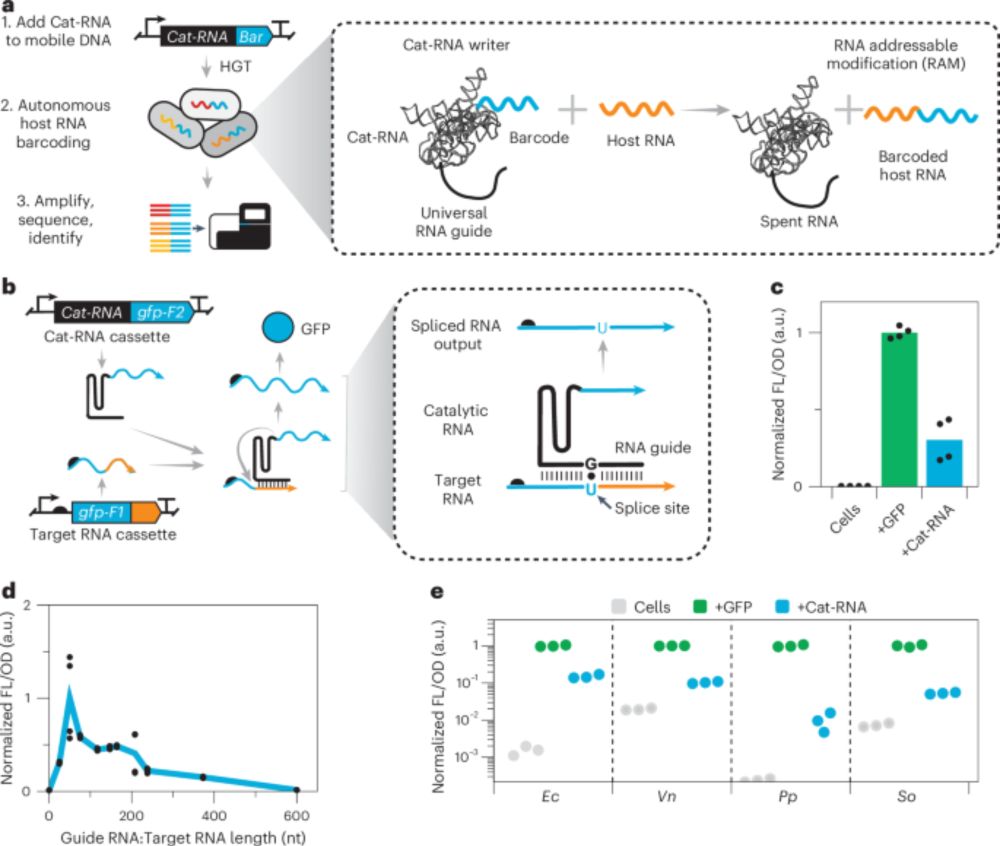
Information storage across a microbial community using universal RNA barcoding - @lauren-stadler.bsky.social @joffsilberg.bsky.social go.nature.com/440gL5E
18.03.2025 18:37 — 👍 15 🔁 4 💬 2 📌 1
New paper out in @bloodjournal.bsky.social. In this study lead by Daria Karpova and Hector Encabo, we investigated the effect of frequent blood donations (FD) on clonal hematopoiesis (CH). Briefly, FD selects for DNMT3A mutations rendering HSC sensitive to stimulation by EPO. doi.org/10.1182/bloo...
12.03.2025 08:03 — 👍 6 🔁 1 💬 0 📌 0
🚀 Join the #ISCO2025 community at @mdc-berlin.bsky.social on May 12-13! 🚀
Don't miss this incredible opportunity to connect with top #singlecell & #spatial omics experts and participate in a special Workshop led by @itaiyanai.bsky.social.
⏳Abstract submissions: March 21)
www.isco-conference.eu
New spatial method, Perturb-FISH, combines imaging-based spatial transcriptomic measurements with CRISPR screening to reveal effects within and between cells. #Science
12.03.2025 17:10 — 👍 17 🔁 4 💬 0 📌 0Want to track clonality in organoids, cancer models, or in vivo transplants? Try our STRACK barcoding libraries from Addgene: www.addgene.org/233208/ www.addgene.org/233210/ www.addgene.org/233209/. If you need help establishing it, reach out—happy to collaborate and troubleshoot
27.02.2025 01:48 — 👍 3 🔁 1 💬 0 📌 0Want to track clonality in organoids, cancer models, or in vivo transplants? Try our STRACK barcoding libraries from Addgene: www.addgene.org/233208/ www.addgene.org/233210/ www.addgene.org/233209/. If you need help establishing it, reach out—happy to collaborate and troubleshoot
27.02.2025 01:48 — 👍 3 🔁 1 💬 0 📌 0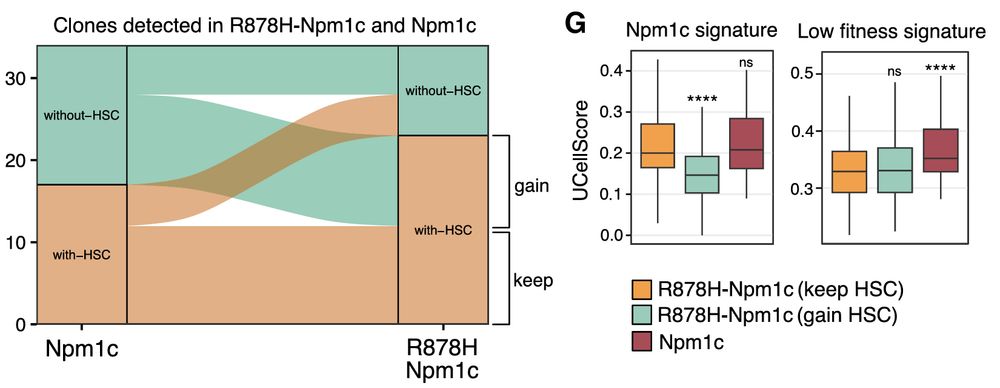
Clonal analysis shows that clones reliant on Dnmt3a for HSC retention are primed to resist Npm1c reprogramming—driving a Gata1-lineage bias. In contrast, clones maintaining HSCs even without Dnmt3a undergo more pronounced Npm1c-driven reprogramming.
27.02.2025 01:48 — 👍 0 🔁 0 💬 1 📌 0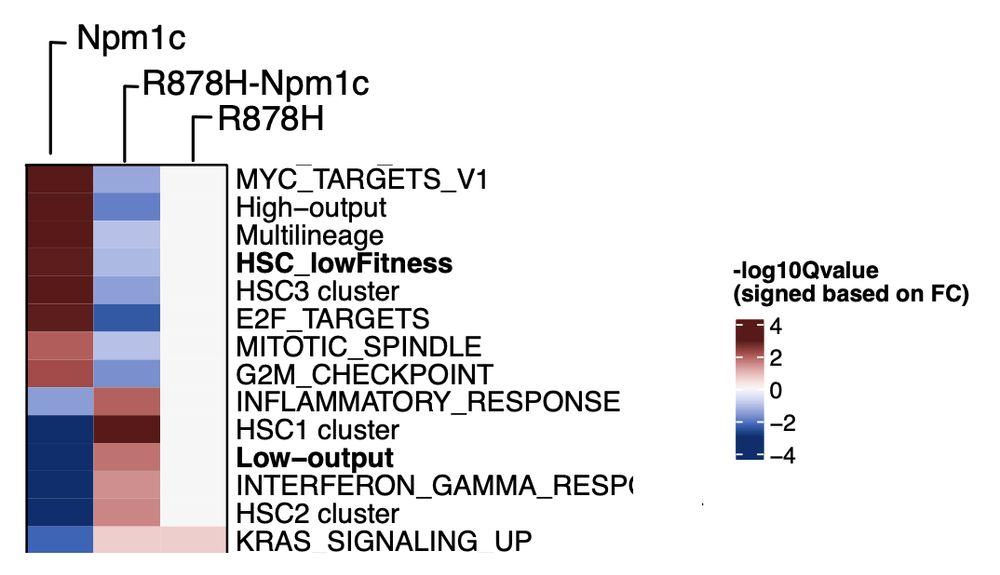
Gene expression analysis shows both Npm1c-only and Dnmt3a/Npm1c HSCs upregulate Npm1c signature genes. However, Npm1c-only cells activate Myc, E2F, and mTOR/PI3K programs, whereas the double mutants suppress these pathways, shifting fate bias.
27.02.2025 01:48 — 👍 0 🔁 0 💬 1 📌 0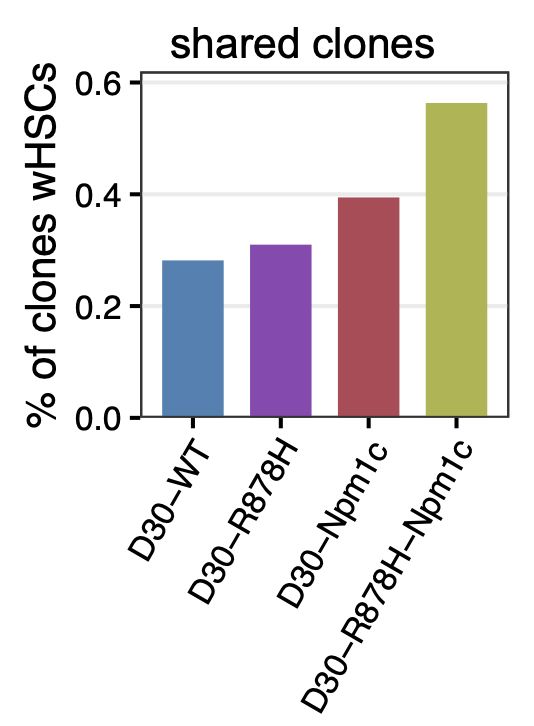
Our sequential mutagenesis experiments reveal that combining Dnmt3a‑R878H and Npm1c mutations produces a synergistic effect—with over 60% of clones retaining HSC identity, far exceeding the outcome of either mutation alone.
27.02.2025 01:48 — 👍 0 🔁 0 💬 1 📌 0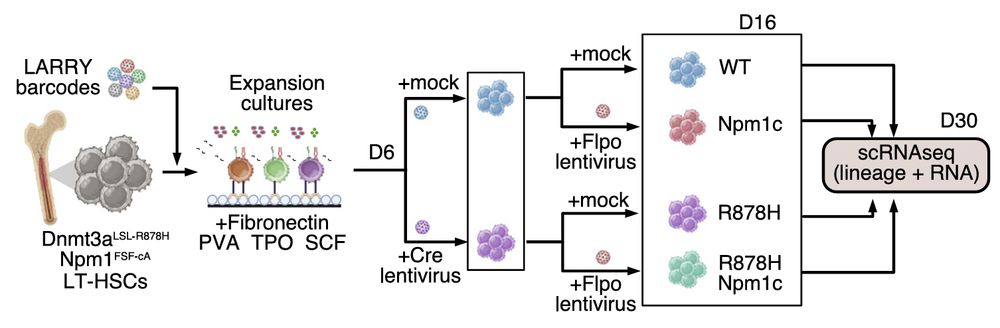
Since most AML cases with Npm1c mutations are preceded by somatic mutations in epigenetic regulators like DNMT3A, we employed STRACK to track stem cell clones and assess mutational synergy
27.02.2025 01:48 — 👍 0 🔁 0 💬 1 📌 0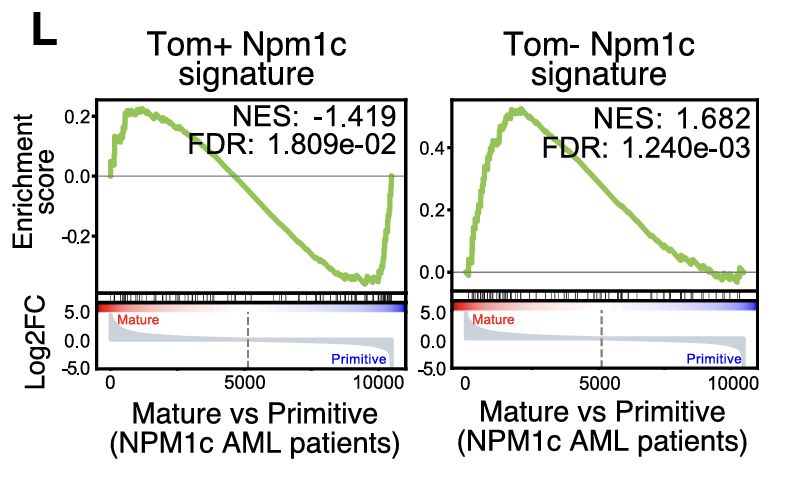
Interestingly, our tdTom+ vs. tdTom– Npm1c gene signatures mirror the mature vs. primitive signatures from patient GSEA—pointing to cell-of-origin as a key driver of heterogeneity in AML!
27.02.2025 01:48 — 👍 0 🔁 0 💬 1 📌 0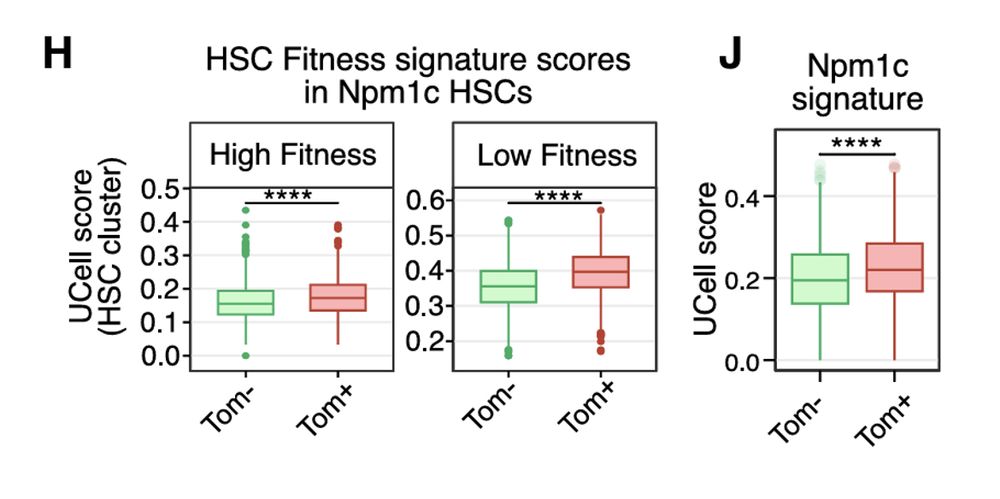
Using above mentioned Flt3-Cre system we then can separate HSCs by intrinsic fitness and found that low-fitness (tdTom⁺) HSCs, when mutated with Npm1c, shift into a primitive, low-output state with heightened Hox/Pbx/Meis activation, in line to our ex-vivo data
27.02.2025 01:48 — 👍 0 🔁 0 💬 1 📌 0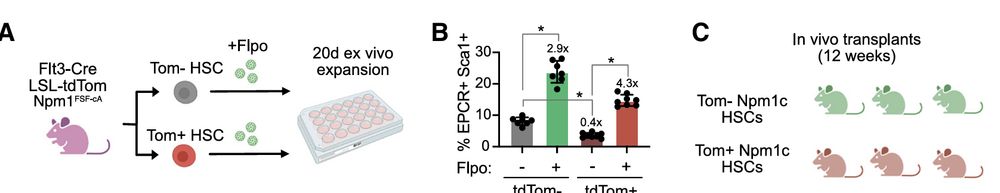
Since Flt3 marks low‐fitness, high‑output HSCs, we crossed Flt3‑Cre; LSL‑tdTom reporter to Npm1c models to validate in vivo, the distinct reprogramming of HSCs sub-compartment by Npm1c mutation
pmc.ncbi.nlm.nih.gov/articles/PMC...
www.nature.com/articles/s41...
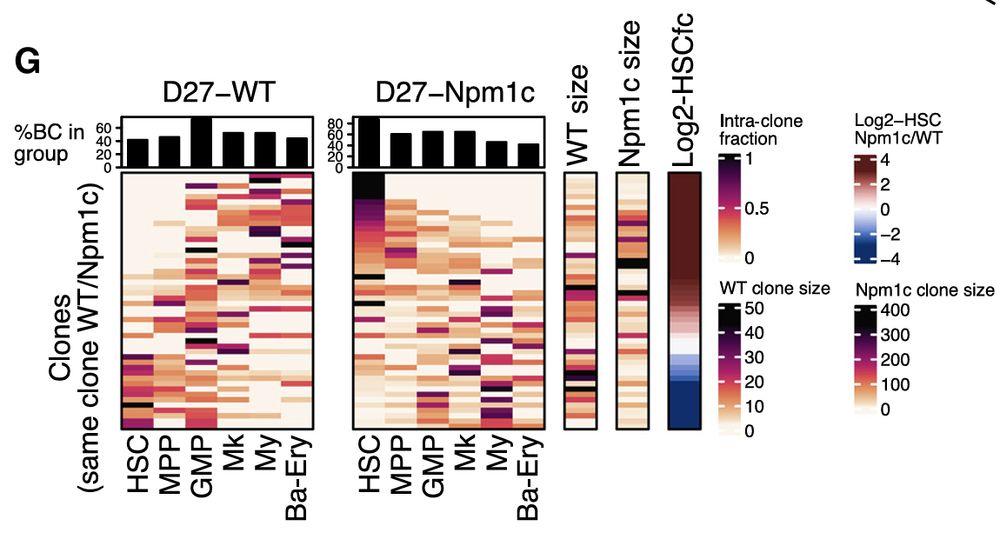
Even more intriguingly, sister clone analysis revealed that Npm1c reprograms HSC fate in a heritable manner: clones with high output in WT become more primitive and differentiation-blocked upon mutation, while low-output clones mature.
27.02.2025 01:48 — 👍 0 🔁 0 💬 1 📌 0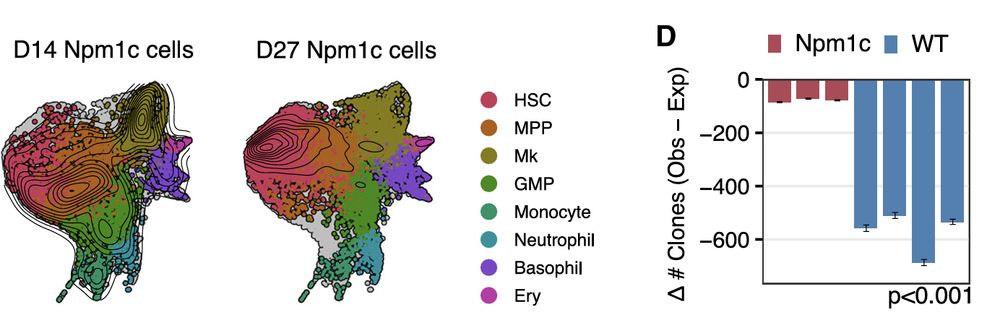
Remarkably, by day 27, Npm1c mutant HSCs not only expanded robustly with near-perfect clonality but also exhibited a striking HSC-like state.
27.02.2025 01:48 — 👍 0 🔁 0 💬 1 📌 0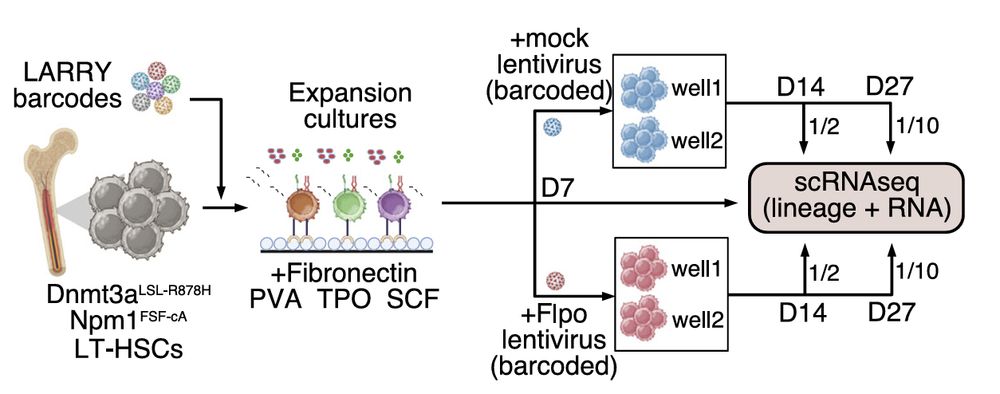
We next wondered if our ex vivo expansion cultures could capture mutation-specific reprogramming. Given that Npm1c is known to massively upregulate stemness program—we set out to test its impact at clonal resolution using our STRACK system.
www.science.org/doi/10.1126/...
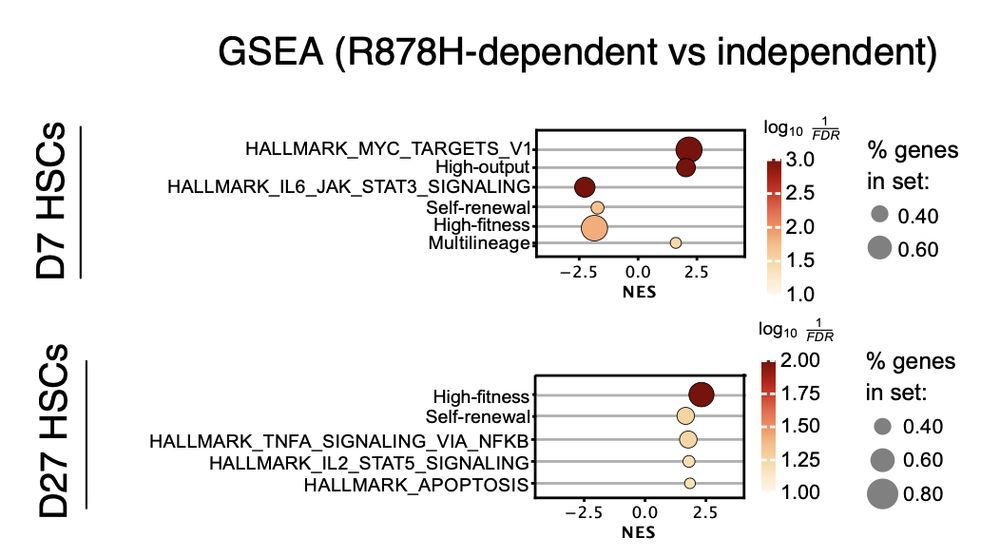
Intriguingly, R878H HSCs and MPPs display reduced expression of early response genes, suggesting dampened inflammatory activation may underlie their competitive expansion.
27.02.2025 01:48 — 👍 0 🔁 0 💬 1 📌 0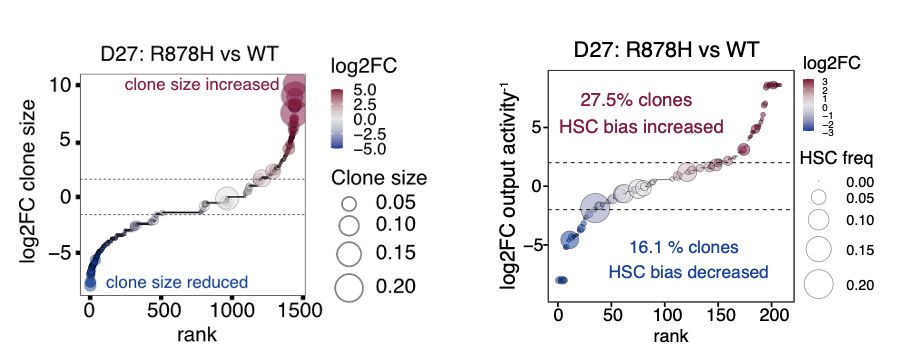
Dnmt3a‑R878H mutant HSC clones expand more robustly than their WT sisters. While overall behaviors appear similar, most R878H clones gain extra HSCs—reprogramming differentiation‑biased cells to favor self‑renewal.
27.02.2025 01:48 — 👍 0 🔁 0 💬 1 📌 0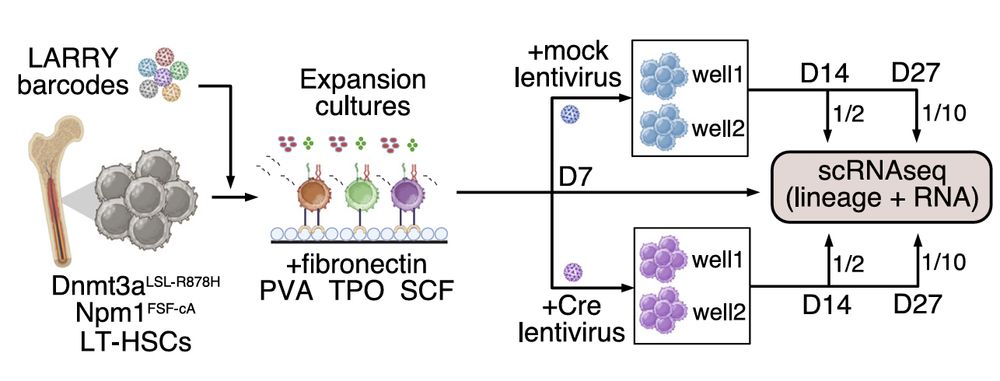
Next, To investigate how different cancer driver mutations influence stem cell fates, we performed STRACK on E-SLAM HSCs from a mouse model carrying conditional knockin of Dnmt3a-R878H mutation (R878H) that can be activated using cre recombinases
27.02.2025 01:48 — 👍 0 🔁 0 💬 1 📌 0