Thanks a lot, Nick!
09.10.2025 09:49 — 👍 0 🔁 0 💬 0 📌 0Pedro Ribeiro
@pedrogribeiro.bsky.social
Postdoc at Petr Nguyen's lab. Lepidoptera genomics | Chromosomal evolution | Phylogenetics Science (and some personal life too) 🇧🇷🇨🇿🦋🍻
@pedrogribeiro.bsky.social
Postdoc at Petr Nguyen's lab. Lepidoptera genomics | Chromosomal evolution | Phylogenetics Science (and some personal life too) 🇧🇷🇨🇿🦋🍻
Thanks a lot, Nick!
09.10.2025 09:49 — 👍 0 🔁 0 💬 0 📌 0Huge thanks also to main collaborators @lepphylo.bsky.social, @joanameier.bsky.social, @kaybiodiversity.bsky.social and many others!
18.09.2025 08:22 — 👍 1 🔁 0 💬 0 📌 0That's it! On Monday I defended my PhD working with the genomics and evolutionary history of the relationship between butterflies and Wolbachia, with a comment on the use of genomics to conservation! Thanks a lot to @pavelmatosm.bsky.social for supervising and mentoring!
18.09.2025 08:20 — 👍 7 🔁 0 💬 2 📌 0
@pavelmatosm.bsky.social has a new PhD position opportunity to work with conservation genomics of European butterflies! The project involves very cool collaborations with @projectpsyche.bsky.social, @10klepgenomes.bsky.social and the LepEUConsortium!
Deadline Sep 12
pavelmatos.wordpress.com/join-us/
This month's ERGA BioGenome Analysis & Applications Seminar will feature a talk by speaker Pío Sierra about a new tool, pantera, which allows faster identification of Transposable Elements in genome assemblies. www.erga-biodiversity.eu/post/faster-...
04.07.2025 06:35 — 👍 22 🔁 9 💬 0 📌 0cool! thanks a lot for the tip, I will try that because so far no luck with that!
30.06.2025 06:46 — 👍 1 🔁 0 💬 1 📌 0Anybody out there with BayesAss2 experience? I think we are not understanding each other about how the input file has to look like!
25.06.2025 09:14 — 👍 0 🔁 0 💬 1 📌 0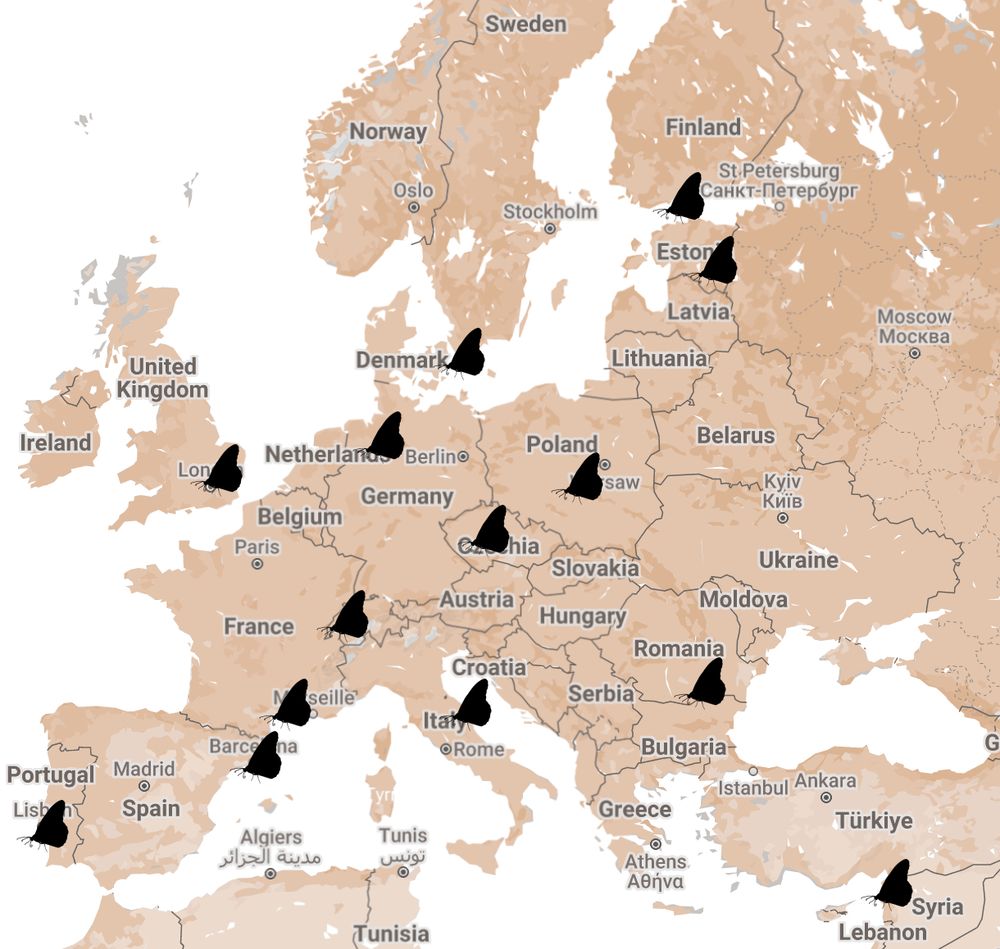
The aim of this proposal is to place population genomic insights into a comparative framework to gain fundamental insights into the determinants of evolutionary outcomes. The project will work within LepEU, the European Lepidopteran Population Genomics Consortium (https://lepeu.github.io/). LepEU provides access to field samples from European populations of diverse species. Chromosome-scale reference genomes are provided by Project Psyche (https://www.projectpsyche.org/). Networking during the postdoc will be facilitated by participation in the 10kLepGenomes COST Action (https://10klepgenomes.eu/). Existing datasets await analysis, while additional samples need DNA extraction and submission for sequencing. Functional validation capability (CRISPR/Cas9 gene manipulations) is also available to test emergent hypotheses of allele-to-phenotype impacts. Personal research interests of the postdoc will be important to determine the exact project, as the project has a generous sequencing budget.

The successful applicant should have a PhD (obtained within 6 years of the application deadline) in a suitable subject area, such as evolutionary biology or population genomics. A strong interest in population genomics, local adaptation, comparative analyses, and experience working with genomic-scale data is essential. The candidate must have a documented publication record demonstrating relevant skills. Experience working with bioinformatic pipelines (e.g., Snakemake), or working with butterflies is welcome but not essential. The net salary is 28,000 SEK/month (~2,430 Euro, not subject to Swedish income tax) and comes directly from the Carl-Trygger Foundation stipend, which is paid out directly to the postdoc. Only PhD candidates acquired outside of the host department can apply. Currently, the lab of Prof. Wheat consists of 3 postdoctoral researchers, while the Dept. of Zoology provides a vibrant and excellent research environment of active, dynamic researchers. Applications should include: i) a succinct description of research interests and experience, detailing your contribution to any relevant publications (max 1 page), ii) why you are the ideal candidate for this position in the lab (max 1 page); iii) a CV including a list of publications, and iv) the name and contact information of two personal references. Applications will be reviewed on a rolling basis with a deadline of 23 August 2025. The project is planned to start on 1 October, but flexibility in the starting date can be provided for a suitable candidate. Please contact Prof. Wheat for additional information.
🚨Postdoc opportunity🚨: LepEU postdoc: comparative population genomics of European scale adaptation in butterflies
2 year, full-time PD in my group, Stockholm Univ.
Applications assed on rolling basis, deadline: 23 August 2025. Planned start 1 Oct.
Details:
christopherwheatlab.wordpress.com

RAxML-NG v2.0 beta is here
github.com/amkozlov/rax...
Features
1. Difficulty prediction
academic.oup.com/mbe/article/...
2. Adaptive search academic.oup.com/mbe/article/...
3. Early Stop
www.biorxiv.org/content/10.1...
4. Model selection
5. Rapid bootstrapping academic.oup.com/mbe/article/...

(1/4) In its first year, #ProjectPsyche has engaged with the research community by creating networks with experts in moths and butterflies 🦋 Members of the project have also trained the research community in #GenomeCuration, a key step in the generation of genomes 🧬
20.03.2025 07:43 — 👍 17 🔁 10 💬 1 📌 1
This Editorial by Brandon S. Gaut and Claudia A. M. Russo provides some nice insight into how #SocietyJournals work, and why it pays for Scientists to publish with #SocietyOwned Journals. 👇
A Message From the Editors-in-Chief
academic.oup.com/mbe/article/...
#PlantScience #SciencePublishing
It's been brilliant to discuss so many aspects of Project Psyche as a community and find ways to make Psyche even better! Big thank you to @lepphylo.bsky.social @lunduniversity.bsky.social for hosting us and COST for making this meeting possible! www.cost.eu/actions/CA23...
08.03.2025 11:14 — 👍 14 🔁 5 💬 2 📌 0
Do you use Darwin Tree of Life (DToL) genomes for your work or grant applications? If so, can you please fill out this form? docs.google.com/forms/d/e/1F... We are gathering info on how DToL genomes are used. It will help us raise money to continue producing publicly available high-quality genomes.
24.02.2025 20:28 — 👍 50 🔁 70 💬 0 📌 3Official petition for using em-dash in scientific publication (is it used?). It feels to me a bit too flamboyant to use it in a paper, but honestly I see it as a stylish alternative to "i.e" or sometimes colons.
24.02.2025 09:01 — 👍 1 🔁 0 💬 0 📌 0I would like to be added! thanks a lot!
21.02.2025 07:49 — 👍 1 🔁 0 💬 1 📌 0
A postdoc opportunity to work on Noctuidae (Lepidoptera) functional genomics with me, @jadrankarota.bsky.social and @chriswheat.bsky.social in Lund, Sweden. We aim to leverage the amazing amount of data being generated by @projectpsyche.bsky.social and DToL projects.
lu.varbi.com/en/what:job/...

https://coursesandconferences.wellcomeconnectingscience.org/event/k-mer-workshop-for-biodiversity-genomics-20250601/
🚨Registration open🚨 1st - 6th of June, me, Katie Jenike, @rayanchikhi.bsky.social and Gene Myers will run a week long workshop on k-mers for biodiversity genomics organised by @connectingscience.bsky.social at @sangerinstitute.bsky.social. Join us & get equipped to wrestle large genomic datasets!
31.01.2025 15:17 — 👍 26 🔁 17 💬 1 📌 1@lepphylo.bsky.social was ahead of me to post about our paper together with Pavel Matos, @joanameier.bsky.social and others!
I will come back to this to talk a bit more about it.
For me, this is the product of amazing collaborations with inspiring people!
@everyjing.bsky.social maaaybe of interest?
06.01.2025 21:11 — 👍 1 🔁 0 💬 1 📌 0
Happy to have been part of this new article on planthopper (Fulgoromorpha) systematics! A great collaboration between Jagiellonian and Lund Universities!
doi.org/10.1111/syen...
a note on writing: if I could, I would definitely re-edit these articles 😅. I like them, but catch a buncha English mistakes, silly "catch" phrases I'd not use today, syntaxes I'd avoid.
Fun to read tho!

In our lab's website, we have a Blog section for more informal articles: our research, "philosophical" discussions from our meetings and journal clubs.
I've contributed to two of these blog posts:
- Are cryptic species real?
- Congresses attended by our [...]
pavelmatos.wordpress.com/category/blog/
Left ex-twitter today (finally). There I tried to maintain a strictly science-based profile.
Here I will try to be more flexible, bringing some readings, movies, music, personal life in general. But the aim is mostly to engage with researchers!
Cheers!