We are back with an exciting seminar on September 8 at 3pm CEST! We’ll hear from Prof. Birthe Kragelund and Maximilian Vieler (@uu.se).
Register to attend:
tinyurl.com/PMCseminar2
Recordings of previous seminars: www.youtube.com/@PMCModularity
15.08.2025 15:35 — 👍 17 🔁 8 💬 0 📌 1

Helmholtz Munich Joins Germany’s Instruct-ERIC Centre for Structural Biology
#HelmholtzMunich’s BNMRZ has joined Germany’s Instruct-ERIC Centre 🔬
Instruct-ERIC provides open access to advanced structural biology technologies. #BNMRZ advances innovation in #NMR methods, drug discovery, and imaging - boosting the German Centre’s capabilities.
👉 Read more: t1p.de/pjtyv
13.08.2025 07:15 — 👍 11 🔁 6 💬 1 📌 0

Seedless: on-the-fly pulse calculation for NMR experiments - Nature Communications
The authors develop a computational tool to design NMR pulses, based on the GRadient Assisted Pulse Engineering approach, that can calculate optimal pulse shapes on the fly during the measurements, to...
Nature Communications (open) Seedless: on-the-fly pulse calculation for NMR experiments, Charles J. Buchanan, Gaurav Bhole, Gogulan Karunanithy, Virginia Casablancas-Antràs, Adeline W. J. Poh, Benjamin G. Davis, Jonathan A. Jones* & Andrew J. Baldwin* www.nature.com/articles/s41... #NMRchat #NMR 🧲
08.08.2025 14:03 — 👍 7 🔁 3 💬 0 📌 0
European Solid-State NMR School
Do you want to learn about all relevant aspects of biomolecular solid-state NMR, from theory to application?
Join the 10th European Solid-State NMR Summer School❗️
🗓️ October 20-24, 2025 | Garching, Germany
📌 Application Deadline: 01.09.2025
www.bio.nat.tum.de/ocb/upcoming...
#NMRchat #BioNMR
05.08.2025 14:46 — 👍 7 🔁 4 💬 0 📌 0

BNMRZ Symposium Ultra-Highfield NMR and 4D Structural Biology – From Mechanisms to Therapies, November 6–7, 2025, TUM Institute for Advanced Study, Garching @ias-tum.bsky.social registration opens soon www.bnmrz.org/4dstructbiol #BNMRZ #NMRchat #NMR 🧲
04.08.2025 20:14 — 👍 10 🔁 4 💬 0 📌 1
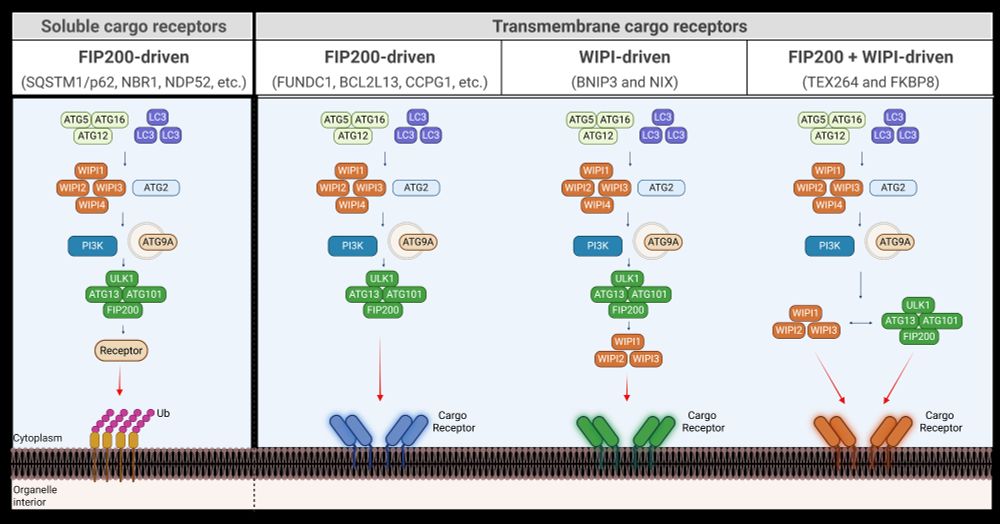
And it's out!
I'm thrilled to share our new paper (Adriaenssens et al., Nat Cell Biol 2025).
This paper describes a new mechanism for the initiation of autophagosome biogenesis.
We found that this WIPI-ATG13-driven pathway is preferentially used by a group of transmembrane autophagy receptors.
25.07.2025 09:43 — 👍 75 🔁 22 💬 6 📌 2
Featuring: The 2026 Biophysical Society Lecture, Lewis E. Kay, University of Toronto, The Essentiality of Solution NMR Spectroscopy in the Post-AlphaFold Era, Monday, February 23, 8:00 PM, Moscone Center www.biophysics.org/2026meeting/... #NMRchat 🧲
03.07.2025 17:08 — 👍 5 🔁 2 💬 0 📌 0
European Solid-State NMR School
We are happy to announce the “10th European solid-state NMR summer school” (October 19-24, 2025) that will take place at the Technical University Munich (@TUM.de) in Garching, Germany. #NMRchat #BioNMR
For more detailed information, please follow the link:
www.bio.nat.tum.de/ocb/upcoming...
11.06.2025 18:37 — 👍 4 🔁 4 💬 0 📌 0

A general substitution matrix for structural phylogenetics.
Abstract. Sequence-based maximum likelihood (ML) phylogenetics is a widely used method for inferring evolutionary relationships, which has illuminated the
New paper from the lab from Sriram Garg in my group. We introduce a general substitution matrix for structural phylogenetics. I think this is a big deal, so read on below if you think deep history is important. academic.oup.com/mbe/advance-...
11.06.2025 14:01 — 👍 95 🔁 52 💬 3 📌 2

Job details
Job details
🚨 We're hiring! Join our excellent Electron Microscopy Facility at @istaresearch.bsky.social as an a Cryo-EM Specialist, providing support for groundbreaking research in a fantastic environment: ist.ac.at/en/job/?titl... #CryoEM #MicroscopyJobs #ScienceCareers
10.06.2025 07:57 — 👍 11 🔁 8 💬 1 📌 1

> Malene R. Jensen @ IBS Grenoble
Research team of CNRS research director Malene Ringkjøbing Jensen at the Institut de Biologie Structurale, Grenoble, France
Two postdoctoral positions in the group of Dr. Malene R. Jensen at the Institute for Structural Biology in Grenoble, France, protein kinase (MAPK) cell signaling pathways (incl NMR, X-ray crystallography and cryo-electron microscopy) www.jensen-nmr.fr #NMRjobs #NMRchat #NMR 🧲
05.06.2025 12:46 — 👍 7 🔁 2 💬 0 📌 0
Nice summary of our ipSAE score for rescoring protein-protein and protein/nucleic-acid interactions in AF2, AF3, and Boltz-1. It works better than ipTM for full-length Uniprot sequences.
03.06.2025 22:50 — 👍 28 🔁 8 💬 0 📌 1
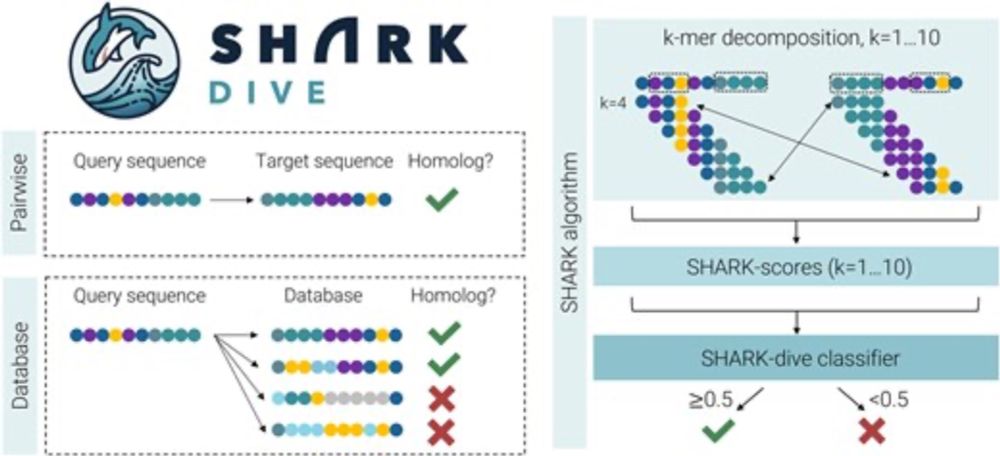
SHARK: web server for alignment-free homology assessment for intrinsically disordered and unalignable protein regions
Abstract. Whereas alignment has been fundamental to sequence-based assessments of protein homology, it is ineffective for intrinsically disordered regions
Are you looking for homologous sequences but alignments do not seem to work? SHARK is available as a webserver: compare sequences or search against precomputed IDR datasets: bio-shark.org @narjournal.bsky.social @mpi-cbg.de @csbdresden.bsky.social @poldresden.bsky.social
doi.org/10.1093/nar/...
28.05.2025 21:24 — 👍 21 🔁 13 💬 2 📌 0
Our manuscript on dsRNA sensor OAS2 from the lab of @oliveiramann.bsky.social is now out in @cp-molcell.bsky.social ! Thanks again to the whole team for their effort putting this story together!
23.05.2025 16:31 — 👍 15 🔁 6 💬 1 📌 0
Still unclear, indeed. I had read a score of 0.7 in your original message but now see that it was 7.0 — sorry about that!
Maybe they reweighted the 0-1 scale?
08.04.2025 20:17 — 👍 0 🔁 0 💬 1 📌 0

Hi Roland, I wonder if this could be the Conservation of Interaction Orientation (CIO) score (0-1) that is mentioned near the end of the Results section? It is derived from geometric analysis of interacting superfamily pairs (ISPs) (Fig 5D, 5E) with a citation to the CIO reference in the methods
08.04.2025 18:26 — 👍 0 🔁 0 💬 1 📌 0
Thank you, Lukas!
11.03.2025 08:33 — 👍 1 🔁 0 💬 0 📌 0
#NMR 🧲
11.03.2025 08:33 — 👍 2 🔁 0 💬 0 📌 0

PhD Position in Structural Biology (f/m/x)
An open #PhD position is available in my lab in Munich 🇩🇪 If you are interested in structural biology, #AlphaFold, or disordered proteins, see below for more details.
jobs.helmholtz-muenchen.de/jobposting/b...
#science #PhD #research
11.03.2025 06:57 — 👍 20 🔁 8 💬 1 📌 1
Congratulations, Woonghee! Very well deserved recognition of your work and your group’s research
26.02.2025 16:03 — 👍 1 🔁 0 💬 0 📌 0

Postdoctoral Research Associate in Disordered Protein Interactions
We're looking for a #postdoc to join our team at KCL Chemistry @kingsnmes.bsky.social. If you're interested in disordered proteins, please get in touch!
3 year post. Ad closes 12 March: www.kcl.ac.uk/jobs/105866-...
24.02.2025 16:57 — 👍 15 🔁 10 💬 0 📌 1
🧵 on our preprint: Zhang & Skolnick's TM score for comparing model of protein to experimental structures of same protein. The d_j are essentially the same as the aligned error in Alphafold. After any structure alignment, it's the displacement of model Calpha from experimental Calpha of residue j.
16.02.2025 02:13 — 👍 60 🔁 16 💬 4 📌 4
Very exciting to see, Gogs! Congratulations and looking forward to seeing Bind develop into the future
17.02.2025 21:55 — 👍 2 🔁 0 💬 0 📌 0
Bind
Bind is a non-profit research organization dedicated to making disordered proteins druggable
NMR will be a cornerstone of our experimental toolkit at Bind. We're developing advanced NMR methods enhanced by AI-based tools to rapidly illuminate disordered protein interactions in powerful new ways. Find out more: bindresearch.org #NMRChat #AI 🧲
11.02.2025 09:49 — 👍 7 🔁 2 💬 1 📌 0
Integrative Structural Biology at the University of Birmingham
Structural & Systems Biology Lab @ University of Geneva | Exploring protein & proteome assemblies | Structured thoughts, intrinsically disordered views
Junior Group Leader @fmp-berlin.de in Berlin - working on integrated #smFRET and #NMR approaches to study intrinsically disordered proteins.
Michael Sattler's Lab @TU_Muenchen & @HelmholtzMunich
We use NMR 🧲 integrated with other techniques to study proteins, RNA & their interactions
Website: https://www.sattlerlab.de//
Protein Disorder in Transcription Lab of Sina Wittmann at @imbmainz.bsky.social
Research Group Leader @chemunicologne.bsky.social at University of Cologne (@unicologne.bsky.social) in Biomolecular #NMR Spectroscopy | We are interested in structural analysis of proteins, peptides and nucleic acids.
https://nmr.chemie.uni-koeln.de/
PhD Candidate, UCSF Biophysics, Kortemme Lab
Computational Biology & AI Lead, Animate Bio
ML for Protein Design
Scientist, synthetic immunologist, loves minimalism. Interested in innate immunity, inflammasomes, cell death, inflammation, autoinflammatory diseases and cancer.
Postdoc in the lab of Carina de Oliveira Mann
TUM, Munich, Germany, interested in pathogens and innate immunity
https://www.bio.nat.tum.de/en/cryoem/home/
Scientist at Boston Children's Hospital, Professor at Harvard Medical School, Husband, Father
Science beyond disciplines — pioneering research in genome biology, immunology, virology & oncology www.genzentrum.lmu.de
Official account of the Gene Center Munich, a central scientific institution @lmumuenchen.bsky.social
Impressum: tinyurl.com/59k5d95t
This is the official account of the Technical University of Munich – Technische Universität München (TUM).
Posts from the web communications team at TUM Corporate Communications Center
Website: http://tum.de/en
Legal notice: http://tum.de/legal-notice
Computational biologist @HelmholtzMunich, prof @TU_Muenchen & associate PI @sangerinstitute. Dad of 4 and mountain lover. Department news, see @CompHealthMuc
Die DFG ist die größte Forschungsförderorganisation und die Selbstverwaltungsorganisation der Wissenschaft in Deutschland.
www.dfg.de
Impressum: https://www.dfg.de/de/service/kontakt/impressum
Social Media-Netiquette: https://www.dfg.de/de/service/presse
AI and tissue clearing-based technologies for health.
Director at Helmholtz Munich, Full professor at LMU Munich, and CEO of Deep Piction (@deeppiction.bsky.social)
We’re🇩🇪🇹🇷🇭🇷🇨🇳🇮🇹🇮🇳🇸🇮🇴🇲🇨🇱🇭🇺🇧🇬🇺🇦🇷🇴🇦🇹🇹🇼🏳🌈
Lab of Petr Broz @ UNIL. Cell death, innate immunology and pathogen defense