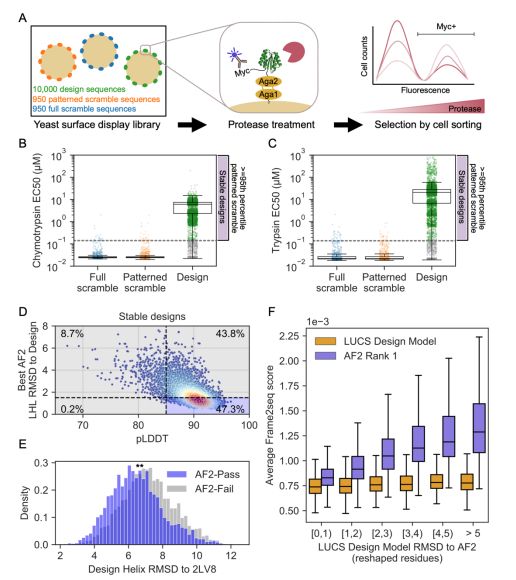
An improved model for prediction of de novo designed proteins with diverse geometries
Fine-tuned AF2 models for "Benjamin Orr*, Stephanie E. Crilly*, Deniz Akpinaroglu, Eleanor Zhu, Michael J. Keiser, Tanja Kortemme. An improved model for prediction of de novo designed proteins with di...
Model weights have been uploaded to zenodo. Fine-tuning and analysis code to be released soon. Work by @benorr.bsky.social, Stephanie Crilly, @dakpinaroglu.bsky.social, Eleanor Zhu, Michael Keiser, and Tanja Kortemme. (9/9) zenodo.org/records/1558...
10.06.2025 18:27 — 👍 2 🔁 0 💬 0 📌 0
This work highlights how augmenting existing models with informative experimental data, as presented here, could expand our exploration of designable protein space and ultimately enable more challenging design problems to be addressed than currently possible. (8/9)
10.06.2025 18:27 — 👍 1 🔁 0 💬 1 📌 0
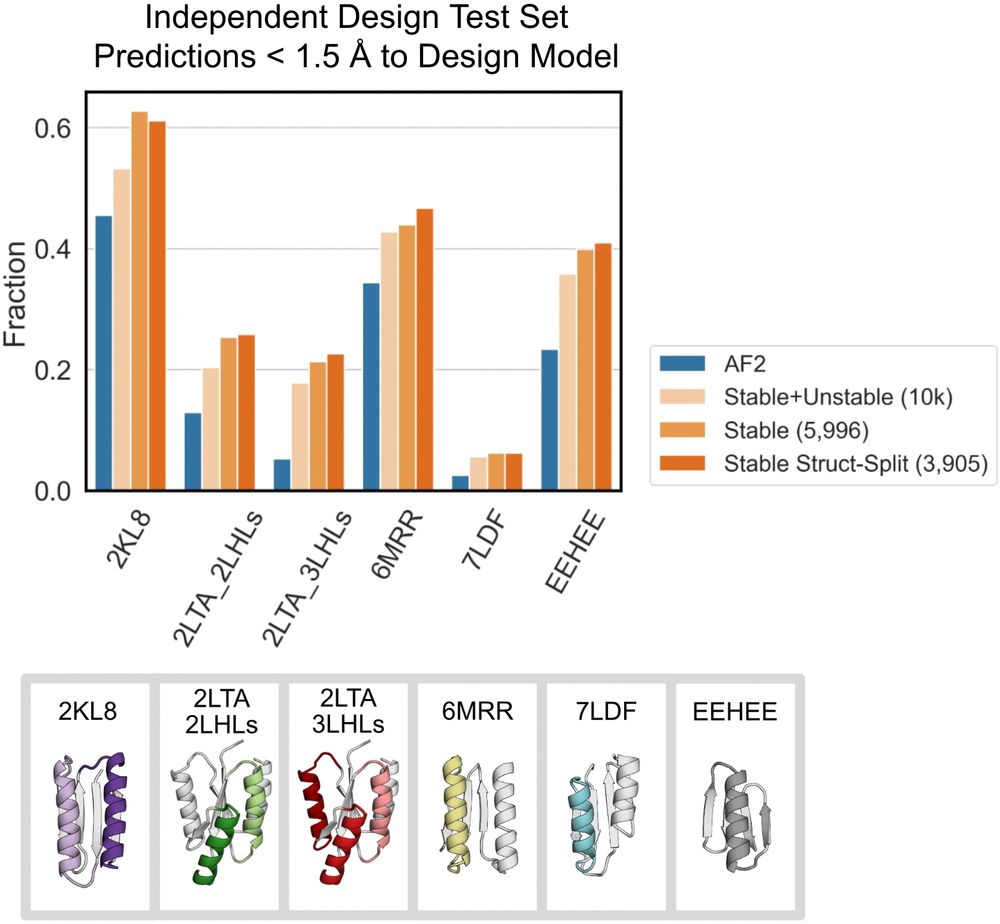
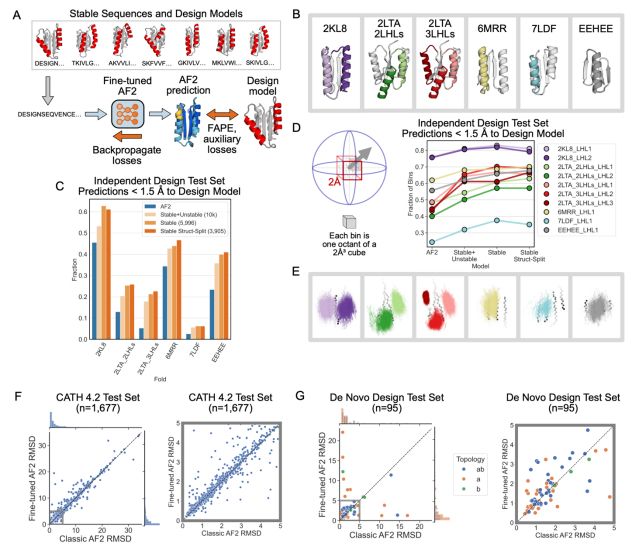
Fine-tuning AF2 on the stable sequences’ Rosetta models improves predictions for geometrically diverse proteins across 5 protein folds. Fine-tuning on ~6k stable designs leads to better performance than fine-tuning on all 10k stable+unstable designs. (7/9)
10.06.2025 18:27 — 👍 0 🔁 0 💬 1 📌 0
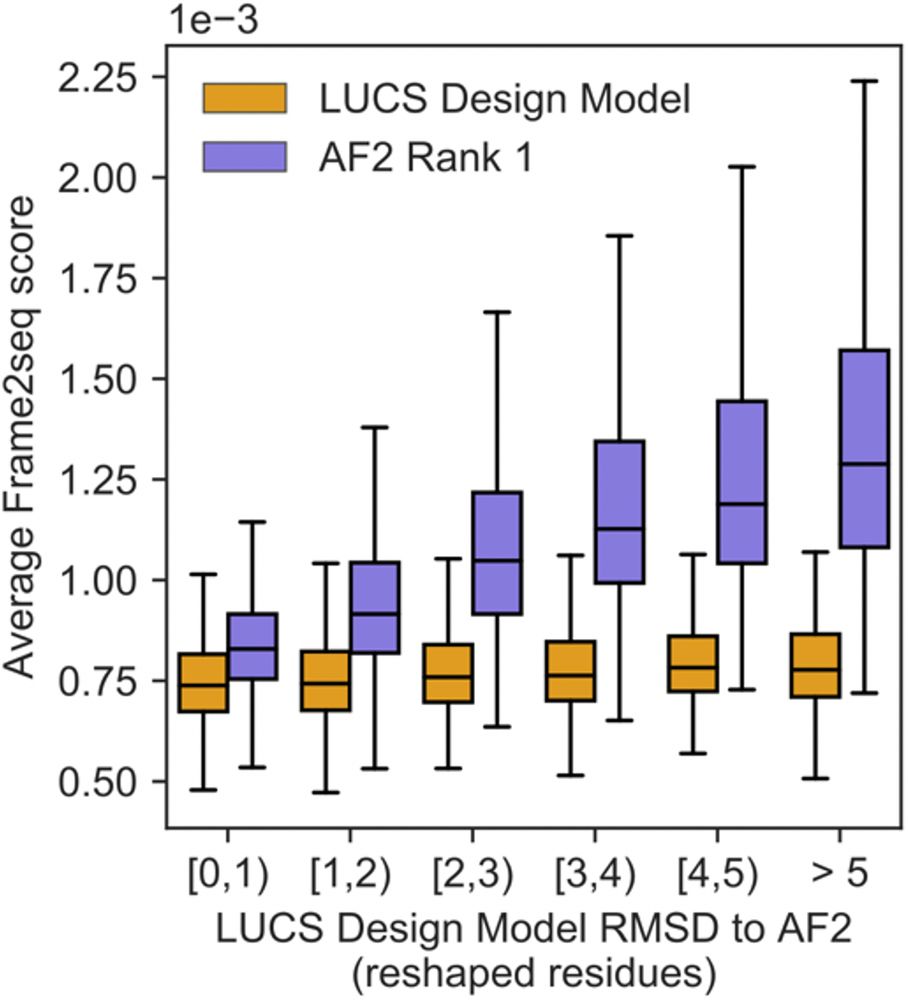
Frame2seq [@dakpinaroglu.bsky.social 2023] scores higher sequence-structure compatibility for the Rosetta models than the AF2 predictions for these stable designs, suggesting that the Rosetta models are more accurate structures than the AF2 predictions for these sequences. (6/9)
10.06.2025 18:27 — 👍 0 🔁 0 💬 1 📌 0
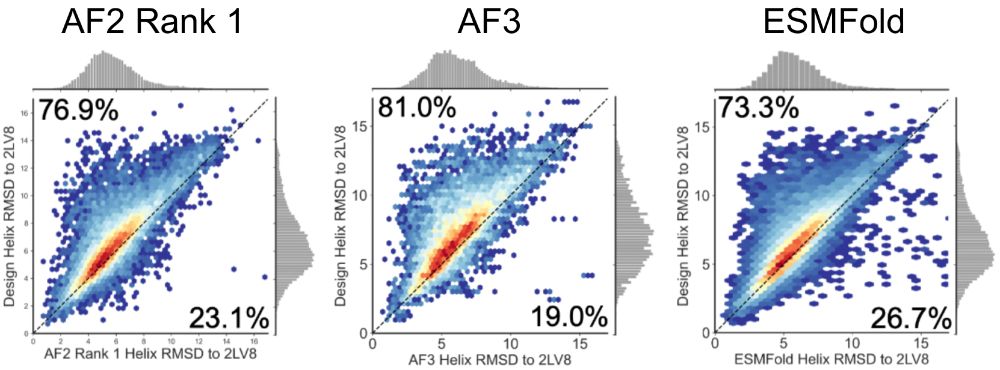
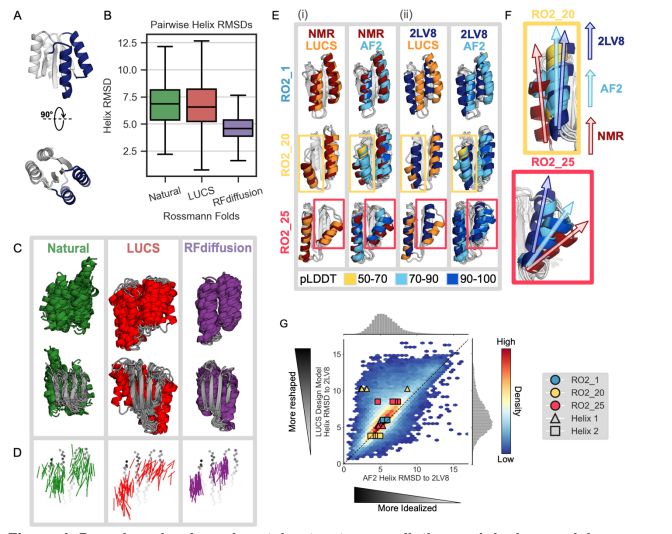
We extended this analysis to 10k diverse Rossmann fold proteins generated by LUCS and tested for stability using yeast display [@grocklin.bsky.social 2017]. For ~6k stable designs, AF2, AF3, and ESMFold all demonstrate a strong bias toward predicting more “idealized” helix geometries. (5/9)
10.06.2025 18:27 — 👍 0 🔁 0 💬 1 📌 0
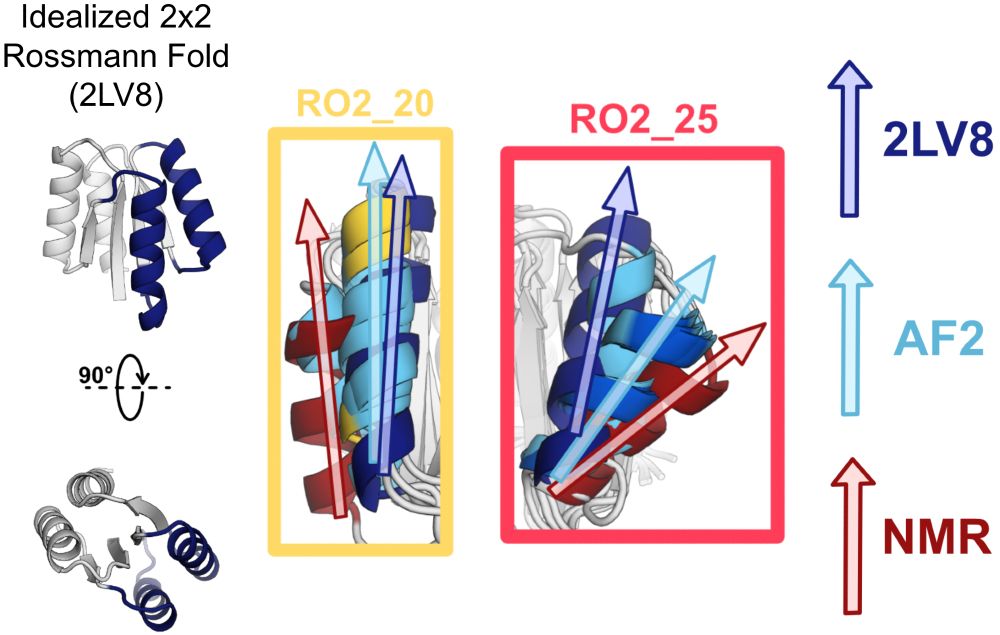
We asked whether protein structure prediction models are biased toward idealized structures for de novo proteins. Indeed, for de novo proteins with diverse geometries, AlphaFold2 predicts structures closer to an idealized de novo protein than the solved NMR structures. (4/9)
10.06.2025 18:27 — 👍 1 🔁 0 💬 1 📌 0
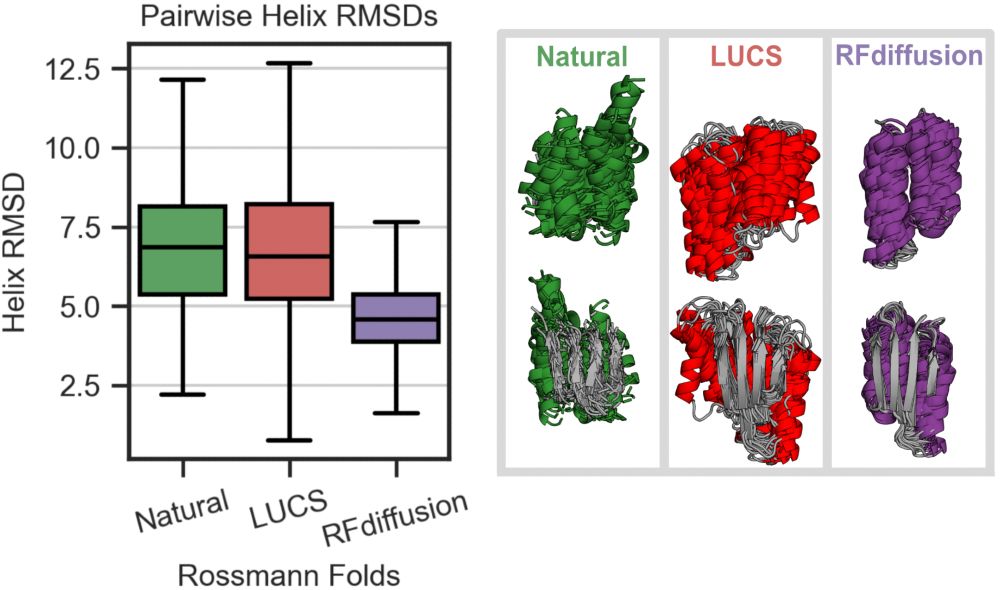
We find that a physics-based method (LUCS) samples greater structural diversity, approaching that observed in natural proteins, in a model protein fold than RFdiffusion, a generative model which utilizes the deep learning-based structure prediction network RoseTTAFold. (3/9)
10.06.2025 18:27 — 👍 0 🔁 0 💬 1 📌 0
In this work we explored how deep learning methods for structure prediction and design may limit our exploration of designable protein space, by favoring “idealized” structures for de novo proteins, and how to overcome these limitations with new data and improved models. (2/9)
10.06.2025 18:27 — 👍 0 🔁 0 💬 1 📌 0
🧬 Structural Bioinformatics | 💊 AI/ML for Drug Discovery | Geometric DL
🔬 @iocbprague.bsky.social, prev. PhD @cusbg.bsky.social @mff.unikarlova.cuni.cz
Data Scientist Generative AI @BayerCropScience. ML for Plant Biology. PhD @IowaStateUniversity https://www.linkedin.com/in/koushik-nagasubramanian/
PhD candidate in Computational Structural Biology @Utrecht University
ML for structural bioinformatics
Computational Structural Biologist @cryocloudio.bsky.social
Bioinformatics Scientist / Next Generation Sequencing, Single Cell and Spatial Biology, Next Generation Proteomics, Liquid Biopsy, SynBio, Compute Acceleration in biotech // http://albertvilella.substack.com
Research Professor | Founder https://www.mindlinkor.com/ | https://github.com/mctrinh | https://x.com/_mctrinh | 🇻🇳 🇰🇷 🇭🇰 🇮🇹 🇺🇸 🇯🇵
Protein Design enthusiast
He/She/They
Group Leader at Helmholtz Munich 🇩🇪 Disordered proteins 🍝, protein dynamics 🏃♂️, and #NMR 🧲
Past: UW 🦡, Oxford 🇬🇧, NIH 🇺🇸, Toronto 🇨🇦
Incoming postdoc @BSC_CNS | machine learning, structure biology and evolution | Prev @UCSF, @UVA | he/him
edraizen.github.io
Translating the Science of Health & Longevity
USD Bio Professor, Drug Developer
📖 Longer content on LinkedIn:
linkedin.com/in/jon-brudvig-phd-9b1830134
📰Sign up for my monthly Substack newsletter, Translation:
jonbrudvig.substack.com
Assistant Professor at Aarhus University, Denmark | biophysical chemistry PhD | into chembio, synbio, protein design, photochem, microscopy | 2018-2022 at Stanford Bio-X | westberglab.com
PhD Student in the Vorobieva Lab in de novo protein design
Professor at UCSF. Molecular/cellular mechanisms of brain disease. Functional Genomics. Diversity & Inclusion. 🏳️🌈 he/him
Core Investigator @ Arc Institute | Associate Professor @ UCSF | {Computational, Systems, Cancer, RNA} biologist | Co-founder @exaibio @vevo_ai
Publishing the best of biotech science and business. Find us on Twitter, Facebook & Instagram. Part of @natureportfolio.nature.com.
Chief Data Scientist U of CA Health・Director, Bakar Computational Health Sci Institute・Distinguished Prof af UCSF・Entrepreneur・He/him・Opinions ⇨ my own
physician-scientist, author, editor
https://www.scripps.edu/faculty/topol/
Ground Truths https://erictopol.substack.com
SUPER AGERS https://www.simonandschuster.com/books/Super-Agers/Eric-Topol/9781668067666
Automated discovery of AI x Bio preprint papers.