YouTube video by Reuters
AI-designed antivenom could offer new hope for snakebite victims | REUTERS
🧪🐍 AI vs venom!
Our Nature paper with David Baker shows AI-designed proteins can block deadly snake toxins. Now featured by Reuters! 🎥
✅ 80–100% survival in mice
💸 Cheap, fast, scalable
🌍 A step toward better antivenoms
📄 www.nature.com/articles/s41...
🎬 youtu.be/gnvEDMEr2mc
31.07.2025 09:34 — 👍 1 🔁 0 💬 0 📌 0
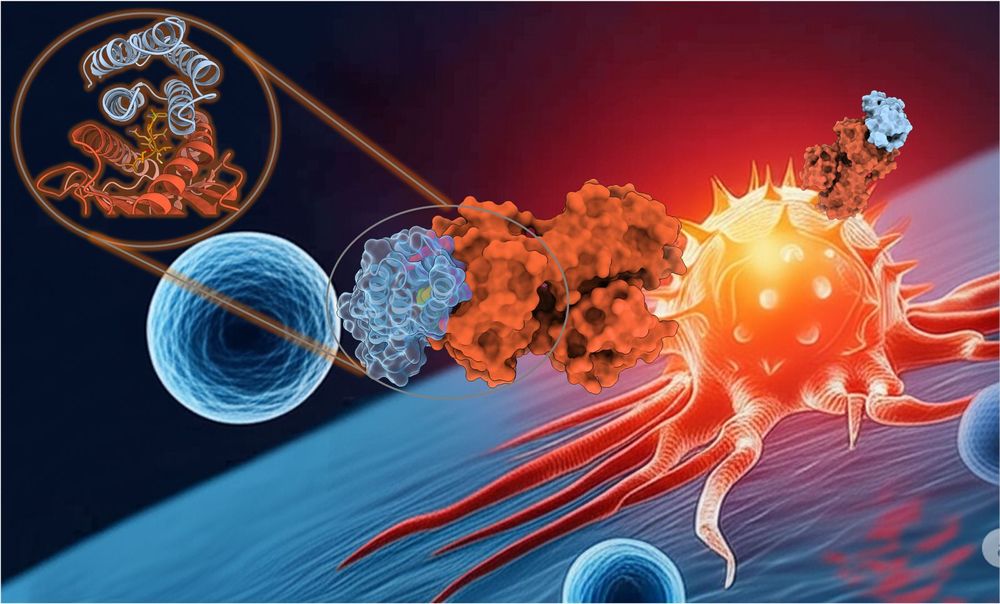
We believe this approach can accelerate the development of safer, more effective immunotherapies for patients.
And a special thank you to the talented @mfernandezquintero.bsky.social & Johannes Loeffler for the stunning graphic! 🎨
#CancerResearch #Immunotherapy #AI
24.07.2025 18:27 — 👍 0 🔁 1 💬 0 📌 0

This was a monumental team effort bringing together immunology, computational biology, and structural biology.
A huge congratulations to the incredible teams at DTU HealthTech, DTU Bioengineering, & @scripps.edu and to all my brilliant co-authors, especially @Kristoffer Haurum Johansen.
24.07.2025 18:27 — 👍 0 🔁 0 💬 1 📌 0

Our platform's key achievements include:
⚡️ Speed: A rapid 4-6 week pipeline from computer design to validated lab result.
🎯 Precision: An AI-driven process to screen for specificity & reduce off-target risks.
🤝 Personalization: It works even for a patient's unique tumor mutation.
24.07.2025 18:27 — 👍 0 🔁 0 💬 1 📌 0
In this work, we designed completely novel proteins, or ‘minibinders,’ from scratch. 🤖
These act as a high-precision guidance system, leading a patient's own T-cells (black) to recognize and destroy cancer cells (red) with remarkable accuracy.
#ProteinDesign #GenerativeAI
24.07.2025 18:27 — 👍 0 🔁 0 💬 1 📌 0
Thrilled to announce our paper, "De novo-designed pMHC binders facilitate T cell-mediated cytotoxicity toward cancer cells," is officially out in @science.org!
We used generative AI to build a 'GPS' for immune cells to hunt cancer.
Read it here: doi.org/10.1126/scie...
24.07.2025 18:27 — 👍 36 🔁 6 💬 1 📌 1
While I’m bouncing between emails and desperately waiting for my holiday, it’s good to know at least something in the lab is holding it together.
Thermal stability might not be glamorous, but it’s underrated.
Designerbodies: even when things get heated, still doing their job ;)
16.07.2025 08:53 — 👍 1 🔁 0 💬 0 📌 0
Built for heat. Unlike me;)
16.07.2025 08:49 — 👍 1 🔁 0 💬 0 📌 0
Designing binders instead of screening for months? Can’t wait to disappoint reviewers in new and innovative ways.
11.06.2025 07:36 — 👍 0 🔁 0 💬 0 📌 0
At AffinityAI we’re moving from “finders keepers” to “designers binders”, one generative model at a time.
If you could have a binder to anything, what would it be?
(Proteins, obscure signalling pathways, that one reviewer who never gets back to you…;)
11.06.2025 07:35 — 👍 1 🔁 0 💬 0 📌 0

Ægget redefined the lounge.
We're redefining what it means to design a protein.
At AffinityAI, we combine Danish design sensibilities with precision molecular engineering. Elegant. Efficient. Engineered to bind.
🧬 Discover how we’re reshaping molecular function:
www.affinityai.bio
02.06.2025 13:49 — 👍 2 🔁 1 💬 0 📌 0
💡 Curious about:
– How generative AI is transforming protein research?
– Why mass spec is such a rich (but messy) playground for AI?
– What it takes to go from spectrum to sequence to structure?
Give it a listen 🎧
30.05.2025 08:34 — 👍 3 🔁 0 💬 0 📌 0

AI giver proteinforskningen turboboost
AI Denmark · Episode
🎧 Had the pleasure of joining Anders Høeg Nissen on the AI Denmark Podcast to talk about how we’re using AI to crack the protein code 🧬
Catch my segment from 9:30 (and yes — it’s in English 😉)
🎙️ open.spotify.com/episode/4g3R...
30.05.2025 08:34 — 👍 2 🔁 0 💬 1 📌 0
Great to see #AffinityAI represented by Esperanza and Oliver at the DTU Startup Day 💪
Lots of interest in what we’re building, especially the fact that we deliver #real #reagents rather than just #AI predictions.
Excited for what's to come!
16.05.2025 11:42 — 👍 1 🔁 0 💬 0 📌 0

🚨 We’re hiring a PhD student! 🚨
Join us at DTU Bioengineering + collaborators Messoud & Håkan Ashina + Esperanza Rivera de Torre to bring preventative migraine therapeutics from 💻 to 🧠
🔗 Apply: lnkd.in/dCRU_EN3
🔍 Project: lnkd.in/dbpi8MwX
14.05.2025 21:33 — 👍 2 🔁 1 💬 0 📌 0
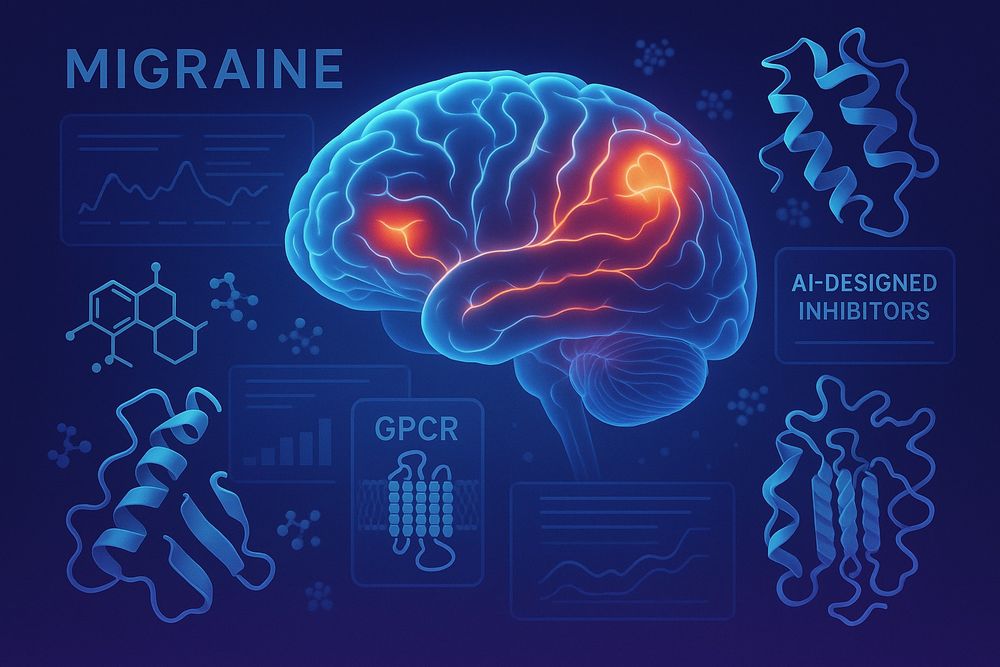
🚨 Big news! We’ve received a NovoNordisk Foundation Tandem Grant for #MIGRAINE — designing #AI-powered #GPCR-targeting biologics for chronic migraine. From computer to human in 4 years. Proud to team up with Messoud and Hakan Ashina, as well as Esperanza Rivera de Torre!
🧠 + 💊 + 🧬 = new hope.
12.05.2025 11:39 — 👍 4 🔁 1 💬 0 📌 0

We’re already supporting:
– Proteins that are hard to purify
– Assays that demand specificity
– Targets that need quantitative detection
Please reach out if you want to hear more.
#StartupLaunch #Founder #AffinityAI
08.05.2025 12:44 — 👍 1 🔁 0 💬 0 📌 0

What we offer:
– Designerbodies™ with pM–nM affinity
– ARCANE™ designs for affinity, stability, and expression
– RAVEN™ screens candidates at scale
Built for diagnostics, therapeutics, and biotech pipelines.
#AIinBiotech #SyntheticBiology
08.05.2025 12:44 — 👍 1 🔁 0 💬 1 📌 0

🚀 Big news: AffinityAI is live
We’ve built a platform for custom protein binders, AI-designed, in vitro validated, and entirely animal-free.
I'm proud to lead this as CEO.
🔗 affinityai.org
#AffinityAI #Biotech #ProteinEngineering
08.05.2025 12:44 — 👍 3 🔁 0 💬 1 📌 0
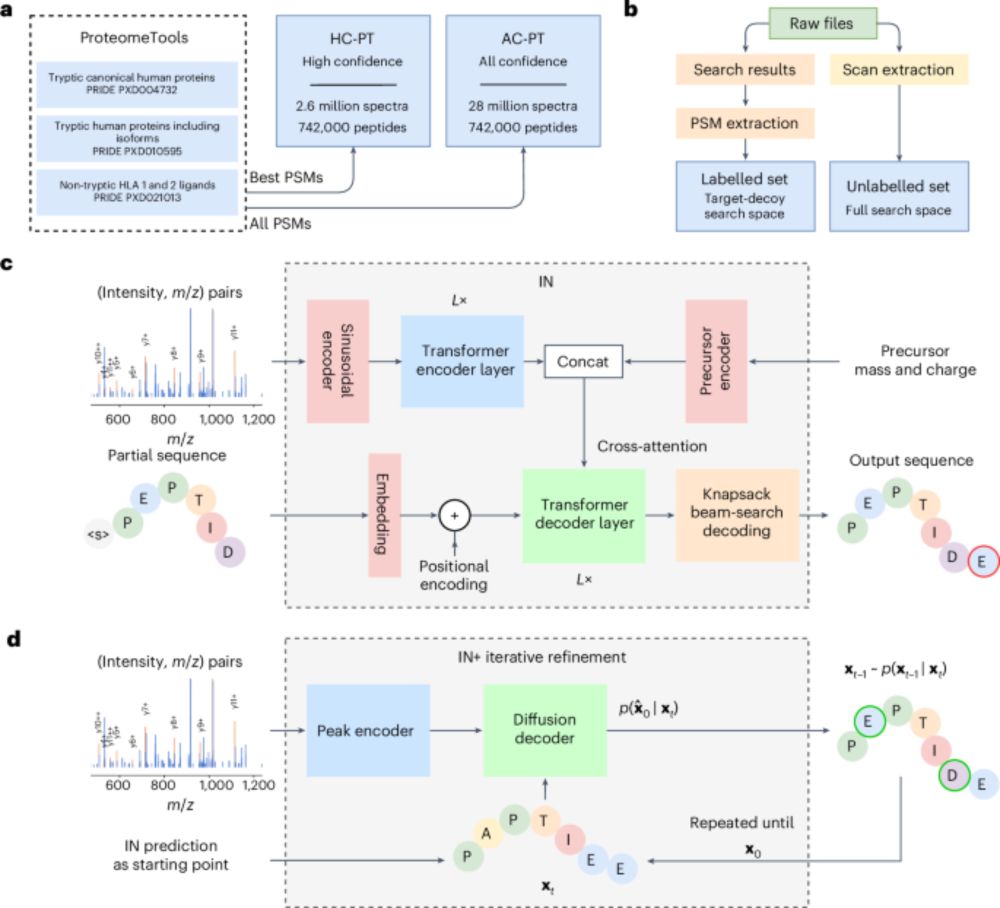
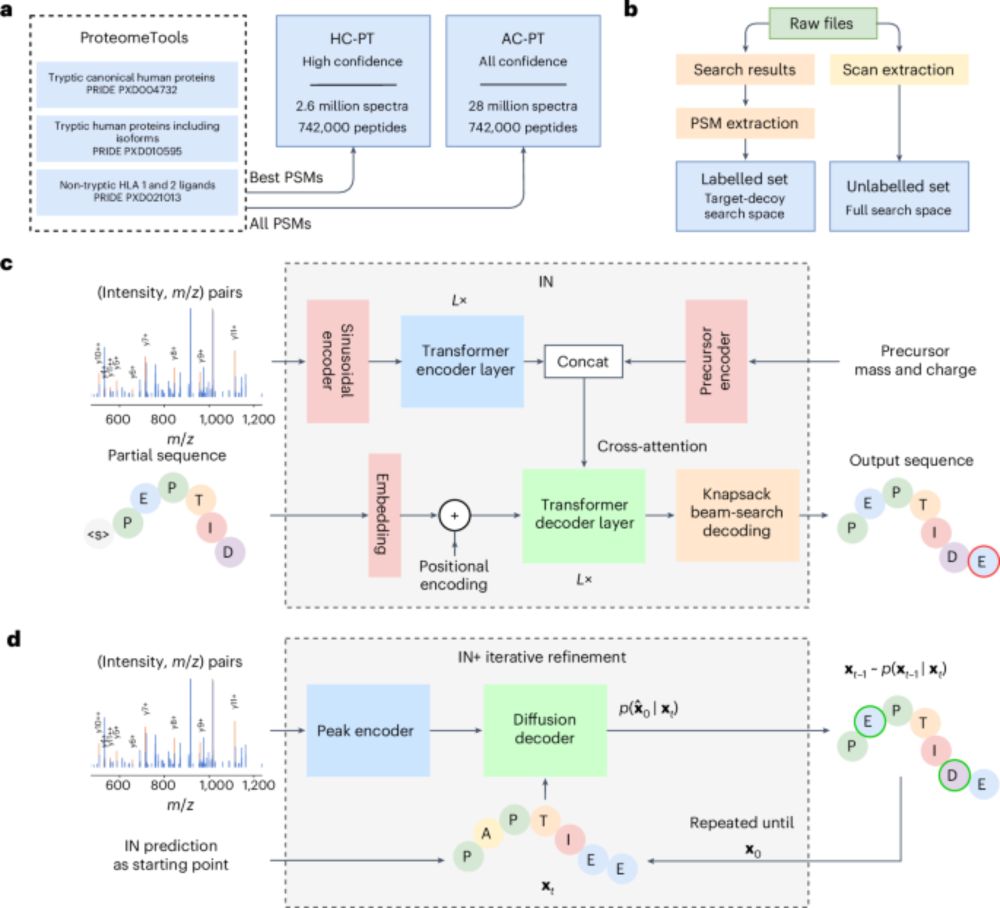
3/
v1.1 generalises well:
In a GluC-treated HeLa dataset, we reached 81.3% peptide recall—higher depth, same model.
Improved confidence metrics = better precision, fewer false positives, more IDs.
Code: bit.ly/43ExRGI
09.04.2025 15:58 — 👍 1 🔁 0 💬 0 📌 0
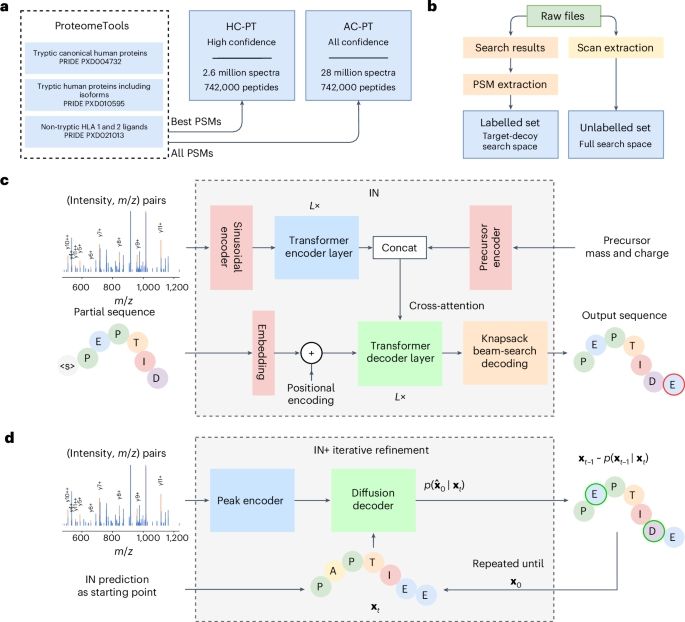
InstaNovo enables diffusion-powered de novo peptide sequencing in large-scale proteomics experiments - Nature Machine Intelligence
InstaNovo, a transformer-based model, and InstaNovo+, a multinomial diffusion model, enhance de novo peptide sequencing, enabling discovery of novel peptides, improved therapeutics sequencing coverage...
2/
What’s new in InstaNovo v1.1?
🧬 +13.5% recall
🎯 +42.6% more exact PSMs
🔍 145% more peptides & 35% more proteins @ 5% FDR
🧪 Support for 7 PTMs: phosphorylation, deamidation, carbamylation, ammonia loss, oxidation, acetylation, carbamidomethylation
Paper: bit.ly/3XBeJFh
09.04.2025 15:58 — 👍 7 🔁 1 💬 1 📌 0

I am very excited to announce the publication of this new research article on "Orally delivered toxin–binding protein protects against diarrhoea in a murine cholera model", which is a topic I sincerely care about: nature.com/articles/s41...
Thanks to all co-authors and funders!
20.03.2025 13:24 — 👍 5 🔁 1 💬 0 📌 0

Excited to share a new paper in Nature Machine Intelligence that I had the pleasure of contributing to, alongside great collaborators from Europe!
InstaNovo is a deep learning model for de novo peptide sequencing — pushing proteomics beyond databases.
📄 www.nature.com/articles/s42...
31.03.2025 09:15 — 👍 8 🔁 3 💬 1 📌 0
Thanks for sharing our work!🎉
01.04.2025 11:41 — 👍 0 🔁 0 💬 0 📌 0
Thanks for sharing our work!🎉
01.04.2025 11:40 — 👍 1 🔁 0 💬 0 📌 0

AI is helping scientists decode previously inscrutable proteins
A new set of artificial intelligence models could make protein sequencing even more powerful for better understanding cell biology and diseases.
New AI tools InstaNovo and InstaNovo+ can now detect proteins previously beyond reach, opening doors in cancer research and disease biology. Early results are promising, though 5% may be false positives. 🧬💻
31.03.2025 15:27 — 👍 14 🔁 3 💬 2 📌 0
Thanks for sharing our work!🎉
01.04.2025 11:40 — 👍 0 🔁 0 💬 1 📌 0
Postdoc @ucl.ac.uk |
Visiting Scientist @crick.ac.uk |
Formerly DPhil @ox.ac.uk @imm.ox.ac.uk
Postdoc at the Technical University of Denmark
Interested in T cells, B cells, antigen recognition, autoimmune diseases and neuroimmunology.
Lundbeckfonden Postdoc Fellow @ DTU 🇩🇰
T cell Immunology | TCRs | high-throughput screens | CRISPR
🔬 Leveraging cutting-edge AI-driven protein design technologies to deliver custom, high-quality affinity reagents at unprecedented speed ⚡🧬
AI/ML for Science & DeepTech | PI of the AI for Materials Lab | Prof. of Physics at UAM. https://bravoabad.substack.com/
Just moved away from "Previously Twitter". It was about time!
#DeepLearning #MachineLearning #AI #LLM #ComputerVision #NLP #NeuralNetwork curated News feed. So dip into the Detphs !
Run by: https://www.linkedin.com/in/eric-feuilleaubois-ph-d-43ab0925/
Sterling Professor of Social and Natural Science at Yale University. Sociologist. Network Scientist. Physician. Author of Apollo's Arrow; Blueprint; Connected; and Death Foretold. Director of the Human Nature Lab: https://humannaturelab.net
🧪 Head of Computational Biology at Protera, based in Paris 🥖
Chief Biological, Clinical and Social Sciences Editor of Nature
Any views expressed here are my own
research engineering @deepmind science | previously #graph #ML for pharma | medicine @ImperialMed engineering @mit_hst
Based at Biomedical and Life Sciences, Lancaster University, UK
We love antibodies, toxins and de novo protein design 🧬🧪🧫💊🖥🐍
Scientist & Assoc Prof in Cell Biology @UFPR
linktr.ee/olgachaim PhD MSc Mol & Cell Bio| Biochemist & Pharmacist
Edits, always original. #OA #TyposON #STEAM #GBM
Alumna @UCSD-MolPharmaco @Unifesp-Biochem @LudwigSP-ExpOnco @UFPR-Pharmacy @EBTG/Danza-Ballet
Postdoc in computational proteomics @CompOmics @VIBLifeSciences - turning public data into tissue biomarkers with ML, AI, and a mission to make proteomics truly reusable
Studying genomics, machine learning, and fruit. My code is like our genomes -- most of it is junk.
Assistant Professor UMass Chan, Board of Directors NumFOCUS
Previously IMP Vienna, Stanford Genetics, UW CSE.
Global health security correspondent @telglobalhealth.bsky.social, based in Bangkok, formerly London.
Contact me: sarah.newey@telegraph.co.uk. Or find me elsewhere - @sneweyy
My work: https://sneweyy.squarespace.com/ / https://linktr.ee/neweysnews
Journalist, presseansvarlig @DTU. tovi@dtu.dk
RE at Instadeep, PhD in computational neuroscience, MSc in CS, interested in ML for life sciences.
Research engineer at InstaDeep working on multi-agent RL