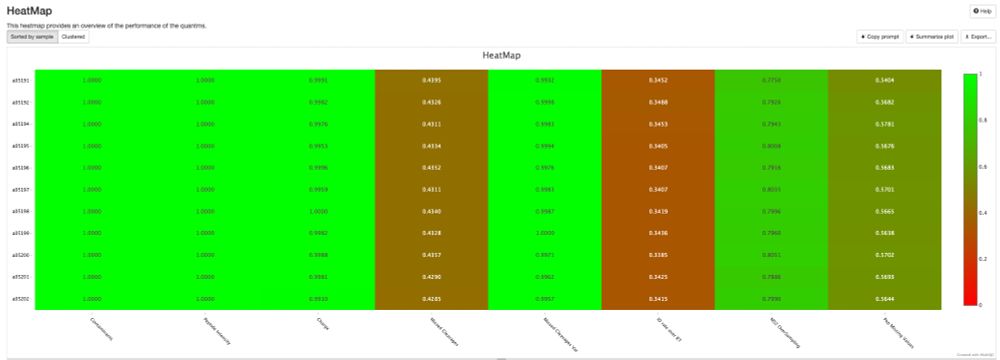
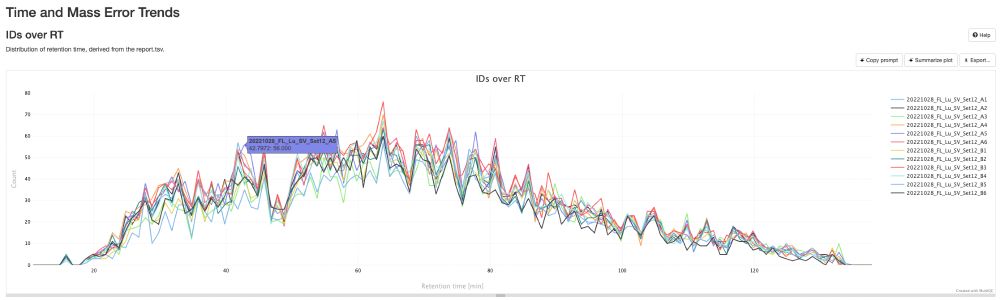
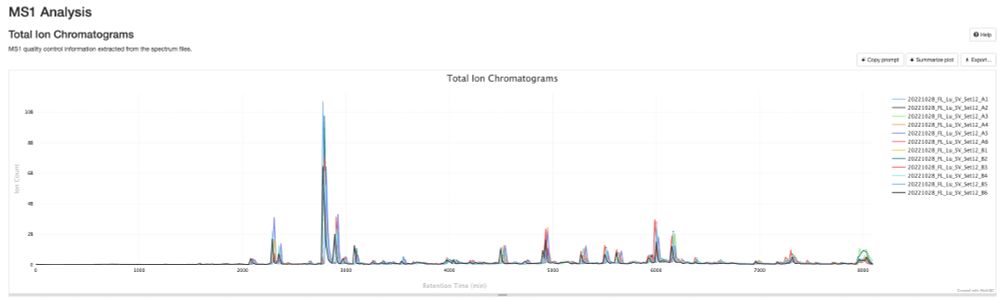
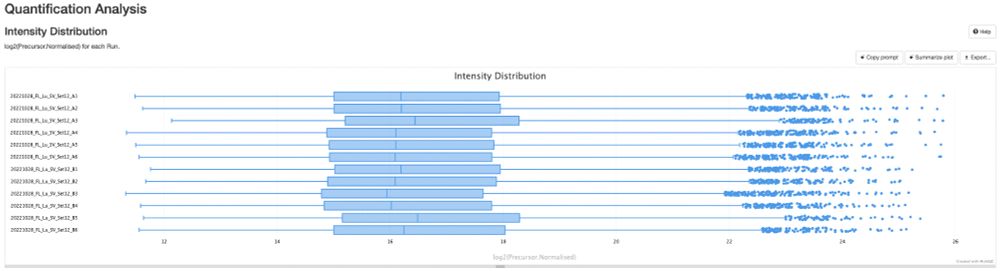
Work of my first PhD student, Sam, who fully stumbled down the de novo rabbithole!
27.08.2025 08:37 — 👍 7 🔁 1 💬 0 📌 0
PSI-AI
Check our website: psi-ai.org
Looking forward to where this will take us!
A huge thanks to all attendees and of course to my co-chairs Samuel Wein and @ralf.gabriels.dev
15.08.2025 10:48 — 👍 1 🔁 0 💬 0 📌 0

Aug14_Kickoff.mp4
Managed Nextcloud - powered by hosting.de
The AI-working group of the Proteomics Standards Initiative officially launched! #AI is increasingly present within #proteomics, we want to support the community to get the most out of their data and models. For this, we need your input #TeamMassSpec
Rewatch the kickoff event: tinyurl.com/37cz9bnj
15.08.2025 10:48 — 👍 10 🔁 4 💬 1 📌 0
Metadata from the paper is often too incomplete to reuse the dataset without extra tools, even raw file species mapping is a disaster! Authors need better reporting guidelines so at least the paper is complete, or they need to make an SDRF (preferably both).
23.07.2025 11:11 — 👍 2 🔁 0 💬 0 📌 0
I fully agree as well, that is the endgoal! That would however require then also treating it as pub with (i) enforcable metadata and peerreview/validation; (ii) doi for citations; (iii) impact metrics for grants.
Maybe the rise of clinical datasets might push this?
22.07.2025 19:31 — 👍 3 🔁 0 💬 0 📌 0
Authors need to inform repositories that their paper got published, right? PRIDE has built-in automatic checks in case the authors would forget. Having a bunch of public datasets that don't link back to a paper is also not ideal. Unless there is proper metadata I'd suppose?
22.07.2025 18:23 — 👍 1 🔁 0 💬 2 📌 0
Never know that there were so many ''condoms'' involved in studying open reading frames, the more you know 🤷♀️
22.07.2025 14:37 — 👍 1 🔁 0 💬 1 📌 0
Yes, enjoyong the amazing british weather, luckily there's tea 😉 I'm flying back directly after the conference but will keep it in mind for future EBI visits 🤗
22.07.2025 10:28 — 👍 1 🔁 0 💬 1 📌 0
Up before sunrise and a little nervous, it’s my first keynote ever!
Speaking tonight at #ISMB2025 in the CompMS track about making proteomics AI ready. Expect metadata, MLMarker, and a lot of public data love. See you there!
@iscb.bsky.social
22.07.2025 05:25 — 👍 6 🔁 0 💬 1 📌 0
ProteomicsML is absolutely awesome to get hands-on with the ML learning part and learn the classical pitfalls
25.06.2025 17:05 — 👍 4 🔁 0 💬 0 📌 0
Depends on where you want to start in your data process (raw files, PSMs, identifications?)
I'd say that AI-readiness is 75% preprocessing and getting all data in the same format plus scalability of your scripts. There are some good packages like psmutils and pyopenMS but depends on your specifics.
25.06.2025 16:00 — 👍 3 🔁 0 💬 1 📌 0
Mark is the cutest octopus which will guide you through #AI driven biomarker identification in your proteomics sample (preprint: www.biorxiv.org/content/10.1...
Pretty clear that @compomics.com is thé best team to do science and have fun with 🙌
23.06.2025 20:03 — 👍 0 🔁 0 💬 0 📌 0
Mark is the cutest octopus which will guide you through #AI driven biomarker identification in your proteomics sample (preprint: www.biorxiv.org/content/10.1...
Pretty clear that @compomics.com is thé best team to do science and have fun with 🙌
23.06.2025 19:58 — 👍 0 🔁 0 💬 0 📌 0

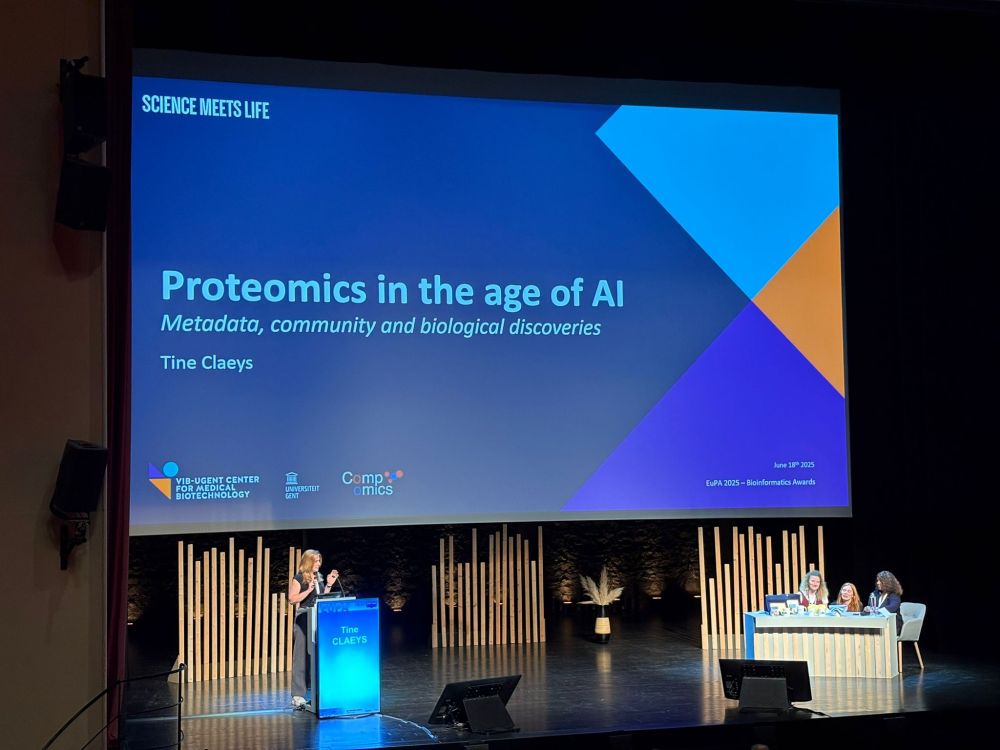
Really liking @tineclaeys.bsky.social's metaphor on metadata:
It's like the philosopher's stone, turning your experimental data into gold and giving it the elixir of life.
Congrats with the EuPA Bioinformatics Award, Tine!
#EuPA2025
18.06.2025 15:06 — 👍 7 🔁 1 💬 1 📌 0
LinkedIn
This link will take you to a page that’s not on LinkedIn
🚨 Has PRIDE helped your research?
Take 15 mins to tell funders why open data matters!
📊 Fill out the EMBL-EBI 2025 survey 👉
www.surveymonkey.com/r/QGFMBH8?ch...
Your feedback helps keep PRIDE open, FAIR & impactful.
🙏 Please share!
#FAIR #OpenData #Proteomics #MassSpectrometry #PRIDE
18.06.2025 06:02 — 👍 10 🔁 12 💬 0 📌 0

And you get the cutest mascotte octopus. His name is Mark! He'll guide you Clippy-wise through your analyses 🙌
16.06.2025 12:32 — 👍 8 🔁 1 💬 2 📌 0
MLMarker is live! This ML-tool predicts tissue similarity and uncovers biomarkers from your proteomics data. It was trained on public data of healthy human tissues.
Preprint & app: www.biorxiv.org/content/10.1...
Let's chat at #EuPA2025 - Award session (Wednesday) & poster session (Thursday)!
16.06.2025 09:31 — 👍 14 🔁 5 💬 1 📌 0
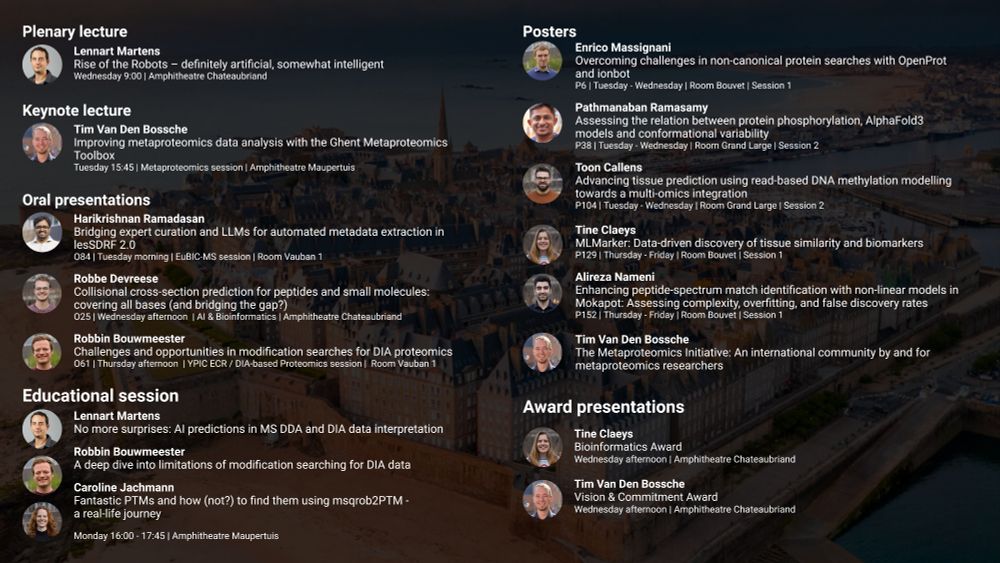
Plenary talk
Lennart Martens
Rise of the Robots – definitely artificial, somewhat intelligent
Keynote lecture
Tim Van Den Bossche
Improving metaproteomics data analysis with the Ghent Metaproteomics Toolbox
Oral presentations
Harikrishnan Ramadasan
Bridging expert curation and LLMs for automated metadata extraction in lesSDRF 2.0
Robbe Devreese
Collisional cross-section prediction for peptides and small molecules: covering all bases (and bridging the gap?)
Robbin Bouwmeester
Challenges and opportunities in modification searches for DIA proteomics
Educational session
Lennart Martens
No more surprises: AI predictions in MS DDA and DIA data interpretation
Robbin Bouwmeester
A deep dive into limitations of modification searching for DIA data
Caroline Jachmann
Fantastic PTMs and how (not?) to find them using msqrob2PTM -
a real-life journey
Poster presentations
Enrico Massignani
Overcoming challenges in non-canonical protein searches with OpenProt and ionbot
Pathmanaban Ramasamy
Assessing the relation between protein phosphorylation, AlphaFold3 models and conformational variability
Toon Callens
Advancing tissue prediction using read-based DNA methylation modelling towards a multi-omics integration
Tine Claeys
MLMarker: Data-driven discovery of tissue similarity and biomarkers
Alireza Nameni
Enhancing peptide-spectrum match identification with non-linear models in Mokapot: Assessing complexity, overfitting, and false discovery rates
Tim Van Den Bossche
The Metaproteomics Initiative: An international community by and for metaproteomics researchers
Award presentations
Tine Claeys
Bioinformatics Award
Tim Van Den Bossche
Vision & Commitment Award
From PTMs to proteins, from metadata to metaproteomics. CompOmics has got you covered at #EuPA2025!
16.06.2025 08:05 — 👍 22 🔁 8 💬 0 📌 0
We are working on an AI based metadata extraction pipeline from papers, supplementary files and mass spectra. Come to @harirmds.bsky.social's talk at #EuPA2025 for the newest and hottest results!
13.06.2025 21:47 — 👍 10 🔁 3 💬 0 📌 0
PRIDE - PRoteomics IDEntifications Database
EMBL-EBI
🎉 Big news from the @pride-ebi.bsky.social Team! 🎉
We’re officially in #ASMS2025 mode and kicking things off with a major milestone:
🚀 Introducing PRIDE-AP – the first Affinity Proteomics archive!
🔗 www.ebi.ac.uk/pride/archiv...
A new home for your #Olink and #SomaScan and other non-MS data.👇
04.06.2025 13:45 — 👍 18 🔁 5 💬 2 📌 0
What an amazing ride these past 2 years with @ypic.bsky.social! So proud of what we have built together.
Now it is your turn! And it is such a great opportunity to support #ECRs and grow with the #proteomics community 🥰
Highly recommend applying! 🙌
03.06.2025 07:58 — 👍 3 🔁 1 💬 0 📌 0

Wondering about the next step in your proteomics career? Curious to know how proteomics experts ended up at their current positions? Looking for tips to help you discover what you really like? Join our Meet-the-Expert session! Wednesday, June 18th | 12:30 - 13:30 Vauban 2 #EuPA2025 #EuPAFPS2025
29.05.2025 07:50 — 👍 10 🔁 8 💬 0 📌 1
What’s new:
Fewer clicks, smoother annotation, ontology browsing and community suggested terms for single-cell proteomics, immunopeptidomics, etc.
20.05.2025 14:25 — 👍 0 🔁 0 💬 0 📌 0
🚧 Refactored lesSDRF; feedback welcome!
I’ve pushed a new version of lesSDRF to make high-throughput SDRF annotation faster and more intuitive.
Github issue: github.com/CompOmics/le...
Temporary app: refactorlessdrf.streamlit.app
I could use some feedback before making this the new lesSDRF 🙏
20.05.2025 14:25 — 👍 5 🔁 2 💬 1 📌 0
No clear guide exists, sounds like sth we should provide!
For accession: PRIDE has a good API but different file formats can require tremendous conversions and parsing.
For submitting: lesSDRF supports up to 500 files. The team from PRIDE has amazing support so I'd contact @ypriverol.bsky.social 🙌
04.05.2025 13:33 — 👍 1 🔁 0 💬 0 📌 0
PAASTA (Palaeoproteomics And Archaeology, Society for Techniques and Advances) is an open, collaborative ECR-driven community for palaeoproteomics researchers!
Doctoral Training - Internationalisation - Professionalisation - Employability - Quality Assurance
Cover foto: © UGent, Christophe Vander Eecken
Lead Bioinformatician in the Stroud Lab @UniMelb Bio21 Institute | Computational Mass Spec Multi-Omics | Chair of ABACBS Diversity, Inclusion and Accessibility Committee
High-quality datasets designed to spark ideas, solve problems, and drive innovation. Fresh data added all the time for your AI projects, research, or curiosity. Let’s turn raw numbers into real impact 🚀
Mass spectrometry/Proteomics scientist developing SRM-MS assays to measure Complement proteins in mouse plasma.
Large-scale Proteomics (clincial & single-cell), mass spectrometry, biostatistics, machine learning, multi-omics.
Empowering Computational Biology, Transforming Global Science!
The International Society for Computational Biology is a scholarly society for advancing understanding of living systems through computation, and communicating scientific advances worldwide.
🇲🇽🇫🇷🇪🇺 Analytical chemist, specialized in #MassSpec #Proteomics. Leading the Immunopeptidomics Platform at HI-TRON Mainz / DKFZ
PhD student at UniFr - Switzerland
Exploring the depths of autophagy through mass spectrometry-based proteomics
PostDoc in Olsen Group and Arachnid wll-being executive manager. 🕷️🕸️ Working on Spatial Proteomics and extracellular vesicles. 🤗
Mass Spec|Proteomics|Chemical Biology
Metabolomics and lipidomics addict... Scientist, sword fighter, martial artist...
Views are my own!
Proteomics | Mass Spectrometry | Reposts are my bookmarks!
Computational Metabolomics, Open Software Development, Open and Reproducible Research, R, Emacs, Bioconductor, Mountains; also on https://fediscience.org/@jorainer
computational proteomics (+multiomics) @ucl.ac.uk, @cruk-cityoflondon.bsky.social, cell biology (PhD @cellbiol-mrclmb.bsky.social), cellular agriculture (@cellag-uk) & science outreach (various!)
https://complexbroadcast.com
Hello there!
Bioinformatician working in Computational Proteomics.
More about me: https://michabirklbauer.me