The Peterson Lab is recruiting – Postdoc to work on mercury methylation in cultured organisms – Peterson Lab at the SFS
I'm recruiting! Looking to hire a postdoc to work on mercury methylation in cultured microorganisms, investigating the metabolic drivers of this important and enigmatic process. See details here: sites.uwm.edu/petersob/202.... Feel free to reach out to me with questions and please pass this along!
04.08.2025 19:00 — 👍 0 🔁 0 💬 0 📌 0

Check it out, and let me know what you think! This represented a major effort from a large team, and I'm proud to see this work out in the world.
30.03.2025 17:39 — 👍 0 🔁 0 💬 1 📌 0

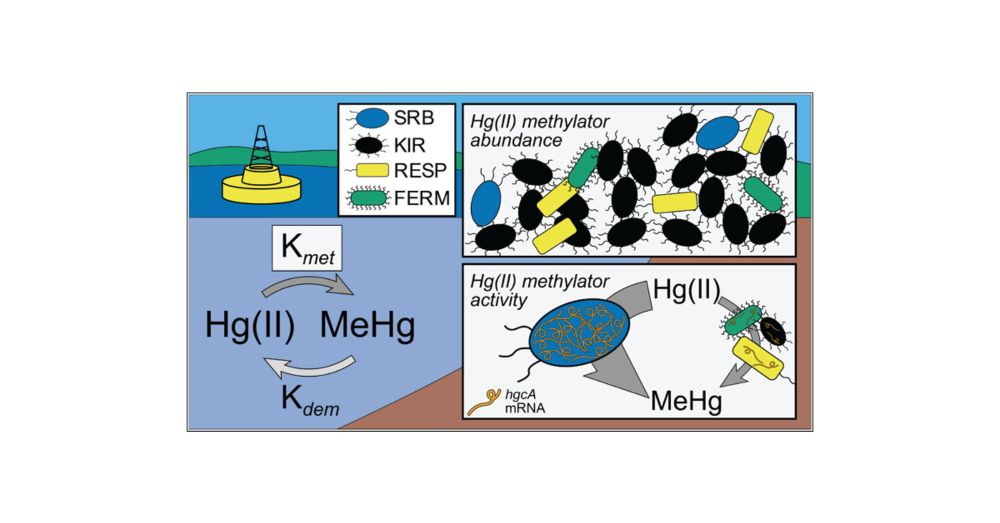
Overall transcriptional activity was also higher in SRB. However, we also identified an arsR-like transcriptional regulator preceding hgcA (previously reported, see text for ref.); these elements were associated with lower hgcA expression and found more frequently in the non-SRB methylators.
30.03.2025 17:39 — 👍 0 🔁 0 💬 1 📌 0

Inhibition of sulfate-reducing bacteria using molybdate resulted in a drastic decrease in MeHg production across all redox conditions. Surprisingly, sulfate-reducing bacteria (SRB) only accounted for <10% of the hgcA gene abundance, but represented the majority (>50%) of the hgcA gene transcription.
30.03.2025 17:39 — 👍 1 🔁 0 💬 1 📌 0
We investigated microbial drivers of methylation using metagenomic and metatranscriptomic sequencing, targeting the mercury-methylating gene hgcA. Gene abundance of hgcA also increased with sulfide, but hgcA expression peaked along with the rate potential, at moderate sulfide concentrations.
30.03.2025 17:39 — 👍 1 🔁 0 💬 1 📌 0
Methylation rate potentials were high, reaching 0.165 day-1. Generally, rate potentials increased with increasing sulfide. Paired methylation and demethylation rates, combined with Hg speciation, strongly suggests that in situ methylation is the primary source of methylmercury in the water column.
30.03.2025 17:39 — 👍 2 🔁 0 💬 1 📌 0

Conceptual diagram of enriched stable isotope incubation assays and supporting data.
We designed a method for conducting enriched stable isotope Hg transformations assays under in situ conditions using custom-designed trace-metal clean bags resuspended in the lake for the duration of the incubation. This was a labor of love... the first trial assays were conducted in 2017!
30.03.2025 17:39 — 👍 1 🔁 0 💬 1 📌 0

Lake Mendota is a well-studied and eutrophic lake with an anoxic hypolimnion for most of the stratified year. Methylmercury concentrations can reach up to 80% (!!!) of total mercury in the hypolimnion, and sulfate reduction appears to be a key pathway driving microbial metabolism.
30.03.2025 17:39 — 👍 0 🔁 0 💬 1 📌 0

First shipment of supplies into the new lab! Can't wait to put these new toys to use.
31.08.2024 04:01 — 👍 0 🔁 0 💬 0 📌 0
As projects develop, I will be looking to recruit graduate students, undergraduates, and postdocs starting in 2025. If you or someone you know is interested in any or all of these topics, stay tuned for these opportunities!
20.08.2024 04:40 — 👍 0 🔁 0 💬 0 📌 0
My lab will focus on contaminant and microbial biogeochemistry, with a specific emphasis on mercury and the Great Lakes region. We will be highly collaborative and use an interdisciplinary approach, with research programs in both environmental and culture-based study systems.
20.08.2024 04:40 — 👍 0 🔁 0 💬 1 📌 0
Professor of environmental engineering at UW-Madison & interim director of Wisconsin Sea Grant. Lots on water chemistry & water quality (plus winter biking in Wisconsin).
Assoc. Prof. @Scripps Institution of Oceanography| UC San Diego
Scripps Center for Oceans and Human Health (SCOHH)
🇺🇸🇲🇱🇦🇿
https://www.aminaschartup.com
https://www.youtube.com/watch?v=-7fIl-lB6-o
IG: schartup_lab
An independent research institute committed to advancing scientific knowledge about ecological systems and their importance to human well-being.
caryinstitute.org
I am a scientist who works (& plays) in waterways to understand their health & function.
Associate Professor in Biological Sciences & Global Change Center Faculty Affiliate @ Virginia Tech. UWYO/Emory alum. she/her. Posts=my opinions. www.hotchkisslab.com
Posts by Shankar Iyer, editor of Trends in Microbiology.
Microbiologist and (meta)genomicist. Picocyanobacteria are my favourite bugs.
NBA, football and tennis enthusiast. Metal, progrock and electronic music variants are my medicine.
We connect science and economics to inform policy that protect, restore, & conserve vital freshwater resources.
https://uwm.edu/centerforwaterpolicy/
I love bacteria.
Biofilm metabolism & antibiotic tolerance dietrichlab.com
Biological Sciences, Columbia University
(a yawn is a silent scream for coffee)
Past, Present and Future of Life
Here and Elsewhere
PI: Betül Kaçar
Account managed by lab members.
Latest paper: https://www.nature.com/articles/s41467-025-67423-y
✨
kacarlab.org
The Conference on Beneficial Microbes. Since 2001, a meeting focus on host-microbe interactions. #BeneficialMicrobesMtg
Our global network is studentsourcing antibiotic discovery through an inspiring and innovative science curriculum. Headquartered at the Wisconsin Institute for Discovery at UW-Madison.
https://TinyEarth.wisc.edu
Professor at the University of Wisconsin - Madison. Researcher on plant-microbe symbioses. Father of 5. Loves hiking, camping, archery, and coffee. Views are my own.
Papa to 9 microbes, mackerel snapper, shepherd, master of sucking. I want to help you learn the skills you'll need to succeed in science.
Professor at UC Davis
Work: phylogenomics & evolvability of host-microbiome systems; #openscience;
Other: #birds; baseball; T1D
Lab phylogenomics.me
Pics jonathaneisen.smugmug.com
Links linktr.ee/jonathaneisen
TED go.ted.com/6WPm
microbiologist, Director of the Wisconsin Institute for Discovery and Professor of Plant Pathology; Chair of the Coalition for the Life Sciences; committed to studying microbial communities and STEM education.
@jo44atWID
Content creator for Elsevier, NPG, and others
Microbiome researcher
Based in Brisbane
Microbiologist turned computational biologist
Bioinformatics Core Director, HPCBio
Open Bioinformatics Foundation Board Member
#metagenomics #microbiome #pangenomics
#python #r (yeah yeah, and #bioperl)
Research and education in modern microbiology at the University of Wisconsin–Madison
NSF-funded network of programs studying ecological processes operating on decadal-to-century time scales. Interdisciplinary researchers and students generate high-impact, team driven science that builds on deep place-based knowledge.