
Dont miss any of our #LongTREC communications at #ISMBECCB2025. Download this flyer to make catching all the latest & hottest long-read transcriptomics research simple.
@anaconesa.bsky.social
@carolinamonzo.bsky.social
PhD. 🏳️🌈 Immunology, aging and bioinformatics enthusiast. @MSCActions postdoc at @i2sysbio.bsky.social @csic.es

Dont miss any of our #LongTREC communications at #ISMBECCB2025. Download this flyer to make catching all the latest & hottest long-read transcriptomics research simple.
@anaconesa.bsky.social

Day 1 of our #SummerSchool : @carolinamonzo.bsky.social , Satrio Wibowo, Mahmud Sami Aydin and @tianyuanliu.bsky.social introducing participants to #LongReads sequencing technologies, how to design #lrRNAseq experiments, library preparation strategies for #lrRNAseq, mapping, QC and more!
14.07.2025 13:24 — 👍 3 🔁 4 💬 0 📌 0We’re kicking off a video series to showcase our network and its mission.
🎥 Watch @anaconesa.bsky.social introducing our project and why raising sustainability awareness in science is so important.
@i2sysbio.es @csic.es
#LongCyclingForScience 🚴🌎
#LongReadsTranscriptomics 🔬 #Outreach4Science ✍
Continuing our LongTREC video series, we’re excited to feature Fabio Zanarello from @crg.eu in this next episode.
#LongReadsTranscriptomics #LongCyclingForScience #SustainableScience
Next in out #LongTREC video series : Nadja Nolte, Doctoral Candidate working with Kristina Gruden and Marko Petek at the National Institute of Biology in Ljubljana, Slovenia. Check the video to learn more about her project!
#LongReadsTranscriptomics #Potato #Poliploidy
Next in the #LongTREC video series, we turn the spotlight to Satrio Wibowo, under the supervision of@minomatt.bsky.social at
@uniofnottingham.bsky.social, Satrio makes the complex ideas of his project easy to understand with his thoughtful explanation. #LongReadsTranscriptomics #AdaptiveSampling
Next Up in the #LongTREC Series 📽️ : Yalan Bi!
Yalan is a LongTREC Doctoral Candidate working with Ralf Herwig at @molgen.mpg.de Yalan shares insights into her research project and her experience as part of the LongTREC network. #LongReadsTranscriptomics #AlternativeSplicing #ProteinInteraction
The LongTREC video series continues with Pablo Angulo Lara, a Doctoral Candidate at @iitalk.bsky.social, where he works under the supervision of Francesco Nicassio.
In this video, Pablo talks about his research and its impact.
#LongReadsTranscriptomics #aneuploidy #cancer #computationalpipelines

📽️ From 🌞 Valencia: Meet Fabian Jetzinger in the LongTREC Series!
Fabian is a Doctoral Candidate at BioBam Bioinformatics, working under the supervision of Stefan Götz, PhD. He is working to make the analysis of long-read RNA sequencing data more user-friendly in #OmicsBox
tinyurl.com/nkszjjm5
📽️ Next in the LongTREC Series: Mahmud Sami Aydin!
Sami is a Doctoral Candidate at @stockholm-uni.bsky.social , working under the supervision of @ksahlin.bsky.social .In this video, Sami shares his research and his role in the broader LongTREC collaboration across Europe.
#AlgorithmDevelopment
🌊 Exploring the Ocean’s Genetic Secrets! Guided by @jm-aury.bsky.social and France Denoeud, @lafucarmen.bsky.social Doctoral Candidate at Genoscope - Centre National de Séquençage, shares how she's using long-reads on marine samples from the Tara Oceans expedition to explore genetic novelty.
13.07.2025 08:37 — 👍 7 🔁 5 💬 0 📌 1


🚀 SQANTI-verse Cards are here! ✨
If you want to get'em all, just follow these steps:
1️⃣ Use one of the SQANTI tools from our Github
2️⃣ Generate amazing data and publish your results
3️⃣ Tag us when you share your publication
We’ll send you a card!
8 are already available & more are coming 🌌
#SQANTIverse

Original article: www.nature.com/articles/s41...
🧬🖥️ 👩🔬 🧪
The article explains how long-read transcriptomics enables the study of isoform expression, discovery of novel RNAs, gene annotation in understudied species, and differential expression between paternal and maternal chromosomes, etc.

I2SysBio @csic.es researchers Ana Conesa @anaconesa.bsky.social, Carolina Monzó @carolinamonzo.bsky.social and Tianyuan Liu @tianyuanliu.bsky.social published an article on #long-read #transcriptomics in Nature Reviews Genomics.
More information: i2sysbio.es/communicatio... ⬇️
📢 Exciting news! We will soon open two PhD positions at @i2sysbio.bsky.social @csic.es, in research projects in collaboration with the @bsc-cns.bsky.social. Both are related to tools to simulate biological tissues. Here’s the description of the open positions. #PhDpositions #PhDOpportunities
👇
Time is running out to register for our #LongTREC summer school - Consider joining us in Norwich this summer at the @earlhaminst.bsky.social to level up your #LongReads #bioinformatics #transcriptomics skills. Click the link below to learn more. We're looking forward to meeting you all in July!
07.05.2025 11:17 — 👍 3 🔁 2 💬 0 📌 0La nota de prensa de @dicv.csic.es de @csic.es resume los grandes logros de @conesalab.bsky.social en I2SysBio. Ana Conesa @anaconesa.bsky.social y Carolina Monzó @carolinamonzo.bsky.social explican su artículo publicado en Nature Reviews Genetics sobre la transcriptómica de lecturas largas #RNAseq
06.05.2025 15:43 — 👍 4 🔁 2 💬 0 📌 0
Científicas del Instituto de Biología Integrativa de Sistemas (I2SysBio), centro mixto del Consejo Superior de Investigaciones Científicas (CSIC) y la Universitat de València, publican una revisión de la tecnología de secuenciación transcriptómica de lectura larga. Esta tecnología permite analizar con más precisión que los métodos tradicionales, las moléculas de ARN presentes en todas las células y que contienen información genética esencial para la vida. Conocer cómo cambia la expresión génica en los diferentes organismos resulta fundamental para comprender mejor el envejecimiento, las enfermedades neurodegenerativas o el cáncer, por ejemplo. El artículo ha sido publicado en la revista Nature Reviews Genetics. La transcriptómica engloba el estudio de todas las moléculas de ARN (ácido ribonucleico) presentes en una célula o tejido. Estas son moléculas intermedias entre el ADN y las proteínas. Si bien el ADN contiene la información genética, el ARN es el que permite la síntesis de proteínas y que la información sea comprendida por las células. Esta rama de la ciencia permite entender qué genes están activos en un momento concreto y cuál es su grado de activación en los diferentes organismos, lo que es clave para comprender el origen y desarrollo de muchas enfermedades humanas.
El @csic.es analiza una nueva técnica de secuenciación génica que ayudará a descifrar enfermedades como el #cáncer, a conocer mejor la resistencia de las plantas o a examinar virus y bacterias. @anaconesa.bsky.social @carolinamonzo.bsky.social
delegacion.comunitatvalenciana.csic.es/el-csic-anal...
Summary of the work of our researchers @carolinamonzo.bsky.social @anaconesa.bsky.social published recently in Nature Reviews Genetics #bioinformatics #transcriptomics #RNAseq 🧬🖥🧪👩🔬
16.04.2025 20:07 — 👍 4 🔁 1 💬 0 📌 0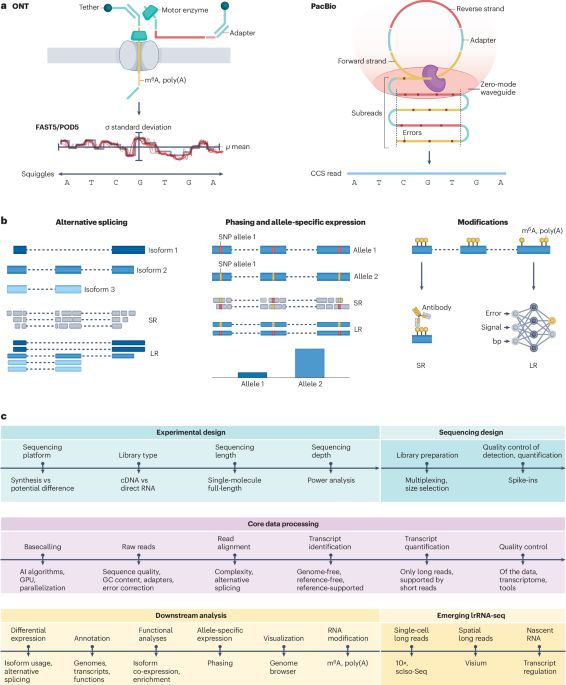
FYI: New online! Transcriptomics in the era of long-read sequencing
18.04.2025 11:24 — 👍 31 🔁 19 💬 1 📌 0
Big thanks to RSG-Spain for their beautiful post summarising some highlights of our recent review on #LongReads transcriptomics published on @natrevgenet.nature.com. bit.ly/lrRNA-seq
@anaconesa.bsky.social @i2sysbio.bsky.social @longtrec.bsky.social

Part 1 and 2 of the Long-read sequencing Special Issues are now live! tinyurl.com/Genome-Res-LRS
14.04.2025 20:13 — 👍 35 🔁 13 💬 0 📌 1Not 1, not 2, but 3 new papers by @conesalab.bsky.social published today in the special issue on long-read sequencing. Find our latest work on SQANTI-reads, genome annotation with lrRNA-seq evidence, and a Perspective in Transcript Divergence. tinyurl.com/GRLong @longtrec.bsky.social
15.04.2025 15:20 — 👍 26 🔁 5 💬 1 📌 2
happy to share our new study! 🧪
while exploring how often #sars-cov-2 "repairs" deletions in BA.1, we found significant genome artifacts in GISAID 🤯 we compared with raw reads from ENA + tested the effects in vitro. turns out, data processing matters a lot 📉
check it out!! doi.org/10.1093/ve/v...

ICYMI: New online! Transcriptomics in the era of long-read sequencing
04.04.2025 11:23 — 👍 11 🔁 4 💬 0 📌 0


It's so incredibly cool to attend a virtual symposium like #SEH2bioinfo! I also loved giving a talk about #TranscriptDivergency 🫶 Thanks for this cool opportunity #RSGspain
02.04.2025 18:36 — 👍 1 🔁 0 💬 0 📌 0Quieres venir a nuestro lab a trabajar conmigo en un proyecto con long-reads transcriptomics? Estás actualmente matriculado en la universidad en el grado o máster? Ofertamos una de las becas de la conexión @bcbhubcsic.bsky.social !!!
02.04.2025 09:31 — 👍 2 🔁 1 💬 0 📌 0I am so happy to see this manuscript finally out!!! We review and discuss all analysis steps in long reads transcriptomics. Hope the community finds this useful! Hugo thanks to @carolinamonzo.bsky.social and @tianyuanliu.bsky.social for the huge work!!! @longtrec.bsky.social @hitseq.bsky.social
28.03.2025 12:52 — 👍 27 🔁 11 💬 2 📌 2mil gràcies!!!
28.03.2025 22:38 — 👍 1 🔁 0 💬 0 📌 0Thank youuu!!! 😀
28.03.2025 22:38 — 👍 0 🔁 0 💬 0 📌 0