We'll have to do a "March for mRNA" 🤦♂️
06.08.2025 00:53 — 👍 6 🔁 3 💬 0 📌 1
A comparison of long-read single-cell transcriptomic approaches https://www.biorxiv.org/content/10.1101/2025.07.03.662955v1
06.07.2025 07:30 — 👍 4 🔁 1 💬 0 📌 0
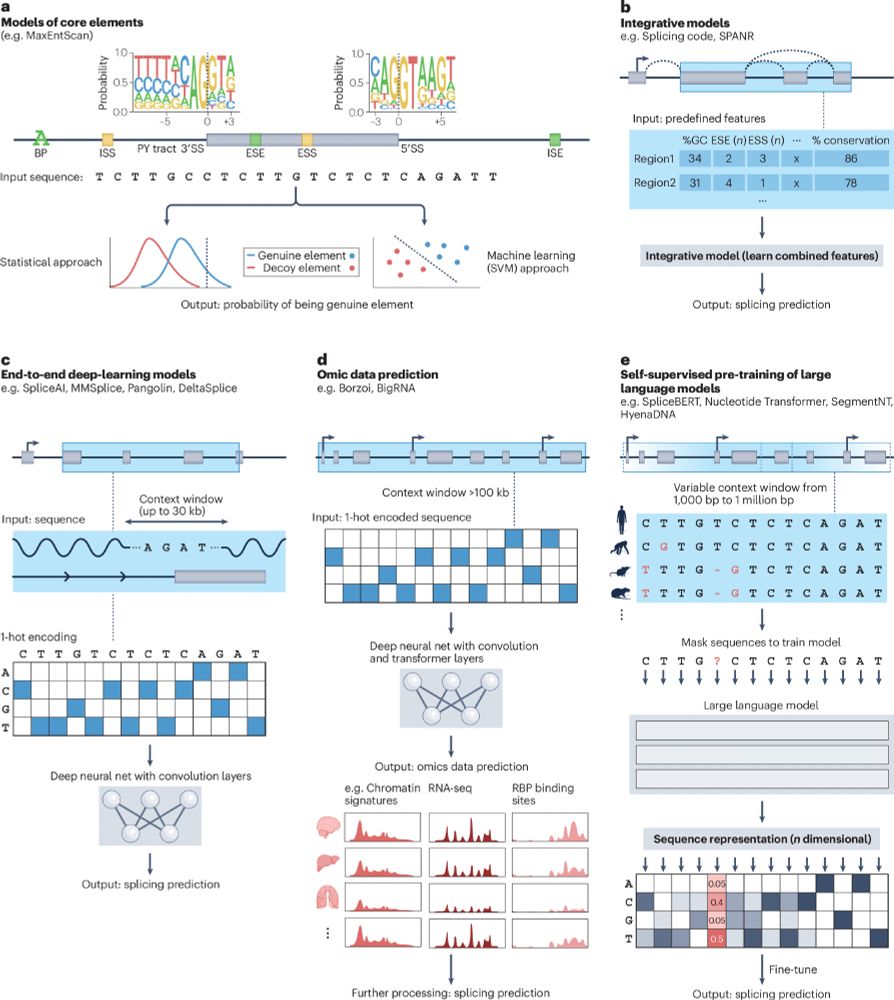
a, Early models built sequence motifs to describe the consensus sequences of individual core splicing elements, such as splice sites (SSs) and intronic and/or exonic enhancers and silencers. Statistical and machine-learning models were built to output the probability of a novel sequence acting as a core splicing element. The sequence logos shown for 5′SS and 3′SS were generated from Human hg38 RefSeq annotations (code available at https://www.github.com/ulelab/splicelogos). b, As our understanding of splicing mechanisms progressed, expert-selected features were extracted from sequences and used to train integrative models to predict splicing outcomes. c, With the advent of deep-learning, models could jointly learn features directly from raw sequence input. Although theoretically, sequence context could be as large as shown in part d, in practice smaller windows of up to 30 kb have been used. d, Supervised models with convolutional and transformer layers produce multimodal genome-wide data. These models use a much larger sequence context and can predict genome-wide data including RNA sequencing coverage, which can be further processed to evaluate splicing. e, By learning how to reconstruct partially masked genomic sequences across multiple species, self-supervised masked language models capture evolutionarily conserved sequence elements and their functional context in a very generic and flexible fashion. The informative numerical representations obtained by large language models can be used for splicing prediction tasks. Here 3′SS within different sequence contexts from multiple species are shown aligned for easier interpretation, but in practice sequences do not have to be aligned. Current masked language models with application to splicing use variable context windows from 1,000 to 1 million base pairs; however, it is currently unclear whether larger context windows confer better performance
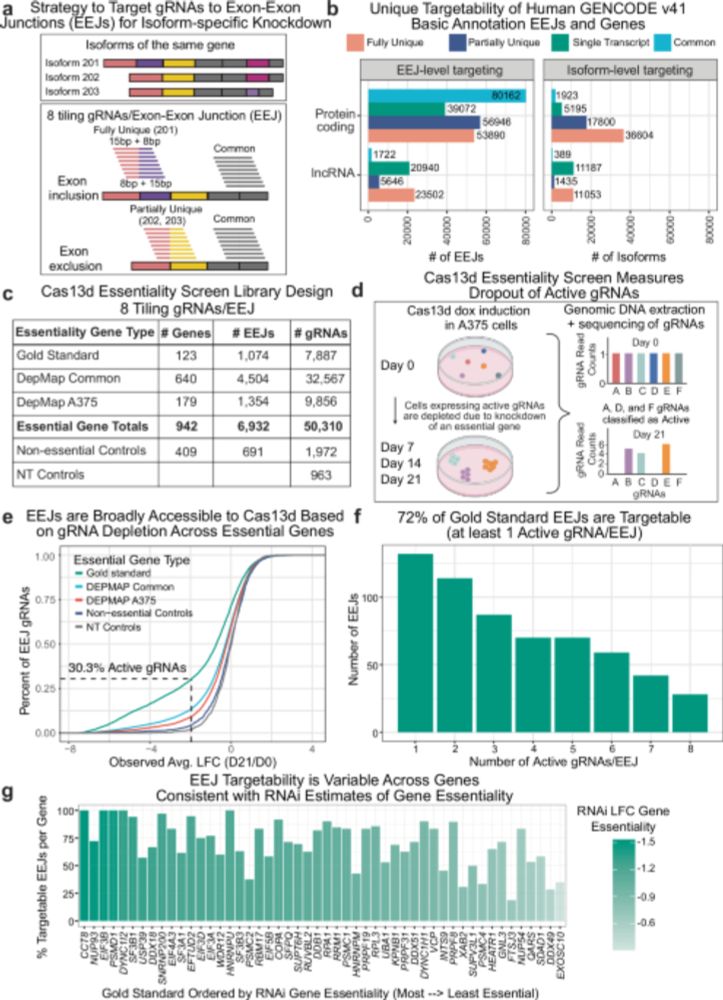
Evolution of splicing model architectures go.nature.com/4eweliE
Figure from our recent Review: From computational models of the splicing code to regulatory mechanisms and therapeutic implications (free to read here: rdcu.be/dVNV4)
02.07.2025 10:21 — 👍 20 🔁 8 💬 0 📌 0

International researchers are fighting for stability in troubled times
We are a group of international scholars at Weill Cornell Medicine (WCM) who moved to New York City to conduct cutting edge research that…
Great day to share this op-ed we co-authored with other international postdocs, highlighting the struggle of doing science under an increasingly hostile political climate, while also fighting for fairer working conditions at WCM: medium.com/@alt_spliced...
19.06.2025 22:34 — 👍 5 🔁 0 💬 0 📌 0
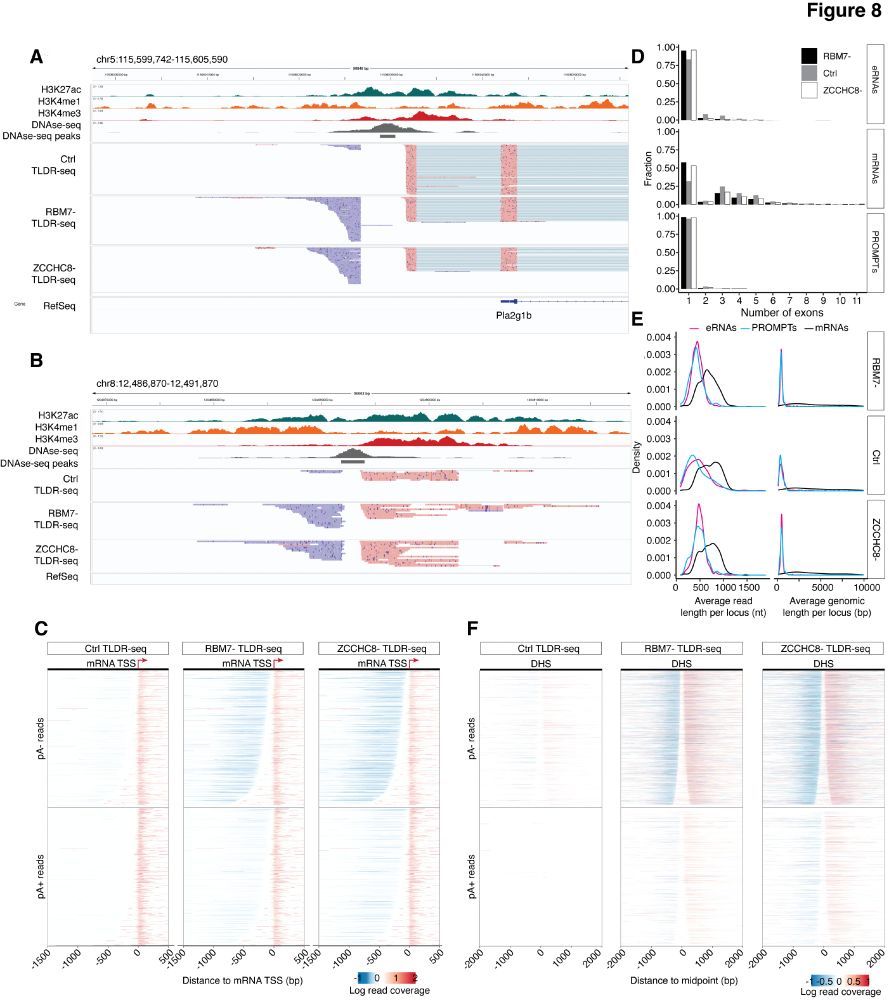
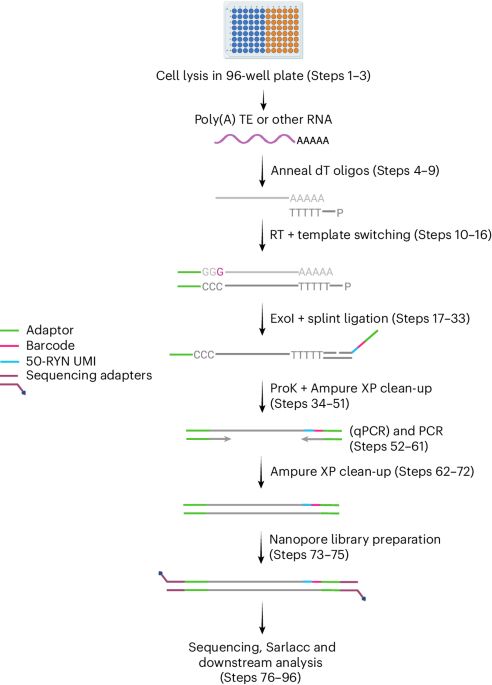
Very proud to share this work just out in NAR: spearheaded by @jamieauxillos.bsky.social and Arnauld Stigliani: TLDR-seq, a method for 5’ to 3’ end long-read sequencing of capped RNAs regardless of 3’ end polyadenylation, based on the @nanoporetech.com platform. (1/4) tinyurl.com/3c2ksdmr
07.04.2025 08:15 — 👍 28 🔁 11 💬 1 📌 1
I am so happy to see this manuscript finally out!!! We review and discuss all analysis steps in long reads transcriptomics. Hope the community finds this useful! Hugo thanks to @carolinamonzo.bsky.social and @tianyuanliu.bsky.social for the huge work!!! @longtrec.bsky.social @hitseq.bsky.social
28.03.2025 12:52 — 👍 27 🔁 11 💬 2 📌 2
Intriguing indeed. The Al'Khafaji lab recently compared PIPseq and 10x for isoform sequencing. Although PIPseq v4, diff. gene capture between platforms is evident. They suggest that some of it might be due to high RNAse content after more cell stress in the PIPseq protocol. bsky.app/profile/aziz...
15.03.2025 16:45 — 👍 1 🔁 0 💬 0 📌 0
With the attacks on science and academia by the current administration, if those of us who have tenure don't speak up, it's really hard to continue justifying the tenure system based on academic freedom.
14.03.2025 17:42 — 👍 1664 🔁 371 💬 22 📌 21
Love this!
26.02.2025 16:17 — 👍 1 🔁 0 💬 0 📌 0
Still a bit more than a week left to apply! PhD opportunity in the Beusch lab. Please share to anyone interested in RNA biology 🧪 #RNAsky #RNAbiology
07.02.2025 17:00 — 👍 10 🔁 15 💬 0 📌 1

Join us to connect with the vibrant #singlecell community.
📢Register for the #ISCO'25 Conference "Innovations in #SingleCell #OMICS" in Berlin!
🗓️ 12-13 May 2025
🎤 Fantastic Keynote and Invited Speakers
🫵🏿 Many slots for talks: submit your abstract
🔗http://isco-conference.eu
Please spread the word!
31.01.2025 10:29 — 👍 14 🔁 9 💬 1 📌 1
VoterVoice - Error 404
Let's hope that the new administration doesn't ban #RNA research from their agenda. In the meantime 2 things: 1) There are some organized actions regarding funding restrictions like form.jotform.com/250226137228... 2) And grants are great but improved postdoc conditions are better #WCMPU-UAW ✊
29.01.2025 03:59 — 👍 3 🔁 0 💬 0 📌 1
I am very thankful to my current and past mentors, amazing colleagues and specially all my peers who took the time to share their insights on the application process.
29.01.2025 03:59 — 👍 1 🔁 0 💬 1 📌 0
I am happy to share that today I got my NOA for the #NCI Early K99/R00! This will support our ongoing efforts to understand RNA splicing impact in cancer phenotypes using single cell multi-omics.
Some good news around all this chaos 💫
29.01.2025 03:59 — 👍 78 🔁 6 💬 5 📌 0
My attempts to use the term "immunospliceosome" in a grant went over like a lead balloon, but maybe "immunoribosome" will gain some traction. This is a SUPER exciting finding, kudos to the authors:
www.cell.com/action/showP...
04.12.2024 19:36 — 👍 10 🔁 3 💬 1 📌 0
Biostatistician @IDEXX formerly at harvardmed, @BIDMChealth, @nasa. Big data, clinical trials, and medical diagnostics. Mainer. Opinions are my own. he/him
Group leader @crg.eu | blood, single cell, synthetic genomics
I study how mutations come about. Based at @columbiauniversity.bsky.social, previously at @normalesup.bsky.social
Father and scientist. Doing dad stuff in Brooklyn and science at New York Genome Center.
𝘈𝘴𝘴𝘰𝘤𝘪𝘢𝘵𝘦 𝘋𝘪𝘳𝘦𝘤𝘵𝘰𝘳, 𝘚𝘱𝘢𝘵𝘪𝘢𝘭 𝘎𝘦𝘯𝘰𝘮𝘪𝘤𝘴
𝘈𝘳𝘤𝘩𝘪𝘷𝘦 𝘰𝘧 𝘮𝘺 𝘵𝘪𝘮𝘦 𝘢𝘵 𝘵𝘩𝘦 𝘰𝘭𝘥 𝘱𝘭𝘢𝘤𝘦 @𝘮𝘢𝘯𝘪𝘮𝘰𝘵𝘢𝘴𝘰𝘭𝘥𝘴𝘵𝘶𝘧𝘧.𝘣𝘴𝘬𝘺.𝘴𝘰𝘤𝘪𝘢𝘭
@𝘮𝘢𝘯𝘪𝘮𝘰𝘵𝘢 𝘰𝘯 𝘮𝘰𝘴𝘵 𝘱𝘭𝘢𝘵𝘧𝘰𝘳𝘮𝘴
Spatial-omics nerd in NYC looking for new ways to connect scientists with the tools they need.
#Trekker
Assistant Professor, Epidemiology at MD Anderson. Molecular epi and statistical genomics to understand cancer and human development
https://bhattacharya-lab.com/
🌱 Environmental Nonprofit & Mega Reuse Center
♻️ Compost • Outreach • Green Jobs
⏰ Open Every Day 10am-6pm (1 12th St. Brooklyn)
Find us at bigreuse.org and instagram.com/bigreuse
linktr.ee/bigreuse
Personal account of the Director of Research at Cure Parkinson's - All views my own - Kiwi - Kia kaha - 侘寂 - https://scienceofparkinsons.com/
Incoming Assistant Professor, Columbia University | @DamonRunyon.org Postdoc, UC Berkeley | PhD Princeton University | Mobile elements, Structural biology and Genome engineering | http://thawanilab.org
Global Alliance for Spatial Technologies: an organized whole that is perceived as more than the sum of its parts. Democratizing Spatial-Omics Techs
Our research group is interested in the molecular and cellular origins and evolution of vertebrate organs. Tweets by Henrik, usually in the name of our group.
home.kaessmannlab.org
Official account of SFB 1531 "Damage control by the stroma-vascular compartment" | Frankfurt am Main | Speaker: Professor Ralf P. Brandes
Website: https://www.SFB1531.de
Director of the Institute of AI for Health @ Helmholtz Munich
PhD Candidate Human Genetics @ University of Chicago & Genscape Lab
https://www.genscape-lab.com/
America/Real Madrid/Yankees/Dolphins fan
Neurobiology - Sapienza University | EMBL
Interests in:
- Visual Systems Neuroscience;
- Neurophysiology and functional neuroanatomy of the Visuomotor System;
- Computational Neuroscience (Modelling & Artificial Neural Networks).
Molecular Cell is a Cell Press journal that aims to publish the best papers in molecular biology.
Molecular and computational biologist. Assistant professor at Boston University in Biology and Computing & Data Sciences. Argentinian
www.fiszbeinlab.com
Associate Professor
DFCI & HMS