Get started by using the notebook on google colab
19.11.2025 14:41 — 👍 0 🔁 0 💬 0 📌 0Vera van Noort
@veravannoort.bsky.social
Professor Computational Biology at KU Leuven and Leiden University Proteins, Peptides, PTMs, microbes, phages, enzymes
@veravannoort.bsky.social
Professor Computational Biology at KU Leuven and Leiden University Proteins, Peptides, PTMs, microbes, phages, enzymes
Get started by using the notebook on google colab
19.11.2025 14:41 — 👍 0 🔁 0 💬 0 📌 0
Code available on github GitHub - sverwimp/AFragmenter: Protein domain segmentation based on PAE values share.google/BpLVdAP8pZ5A...
19.11.2025 14:40 — 👍 0 🔁 0 💬 1 📌 0Want control over segmentation of your protein into domains? Now out in #bioinformatics AFragmenter academic.oup.com/bioinformati... @sverwimp.bsky.social @roblavigne.bsky.social
19.11.2025 07:09 — 👍 3 🔁 1 💬 1 📌 0
Fold first, ask later: structure-informed function annotation of Pseudomonas phage proteins
Structural predictions to improve #phage genome annotation!
@hannelorelongin.bsky.social @gbouras13.bsky.social y.social @susiegriggo.bsky.social @veravannoort.bsky.social
www.biorxiv.org/content/10.1...

Very excited that our work describing hu.MAP3.0 is published in @molsystbiol.org. Here we use machine learning to integrate >25k mass spectrometry experiments to place ~70% of human proteins into 15k protein complexes.
www.embopress.org/doi/full/10....
Looking for a new colleague at the faculty of bioscience engineering www.biw.kuleuven.be/m2s/cmpg/job...
21.05.2025 13:53 — 👍 1 🔁 1 💬 0 📌 0Kregen andere promotoren ook bericht dat ze hun aanbeveling bij @fwovlaanderen.bsky.social niet hadden ingediend terwijl ze dit wel hadden gedaan? #onnodigestress
16.05.2025 08:19 — 👍 0 🔁 0 💬 0 📌 0Zelf nooit hulp gehad, ouders vroegen zelfs niet of ik huiswerk had, ook niet naar resultaten. Nu moeten wij wel helpen, leerstof en opdrachten die mee naar huis komen zijn vaak zo complex dat zelfs wij er moeite mee hebben (2 ouders met PhD).
04.03.2025 09:02 — 👍 1 🔁 0 💬 0 📌 0Check out our preprint on optimal growth temperature prediction #machinelearning #ai #bioprospecting #extremophiles
04.03.2025 06:27 — 👍 4 🔁 1 💬 0 📌 0Would love to read, the pdf on biorxiv seems to be corrupted.
19.12.2024 11:47 — 👍 1 🔁 0 💬 1 📌 0A new #StarterPack with scientist specialized in microbial bioinformatics - #MicroBioInf.
Please comment if you would like to be added: go.bsky.app/5h7VaGm
Looking forward to the discussions!



Lively discussions and live demos of new bioinformatics tools from multi-omics integration to protein structure visualization
17.12.2024 10:24 — 👍 2 🔁 0 💬 0 📌 0
Positive vibes among the master students presenting results of their hard work this semester at the poster session of the Integrated Bioinformatics Project at KU Leuven.
17.12.2024 10:11 — 👍 6 🔁 0 💬 2 📌 0Fast and scalable pangenome inference, core genome inference and subsampling to representative genomes. #comparativegenomics #microbiology applied to over 31,000 lactobacillales genomes!
17.12.2024 08:46 — 👍 1 🔁 0 💬 0 📌 0
Early xmas present, now out in Bioinformatics. SCARAP developed by Stijn Wittouck in collaboration with Sarah Lebeer. PanGenome analysis brought to your laptop academic.oup.com/bioinformati...
17.12.2024 08:42 — 👍 9 🔁 0 💬 1 📌 0Structure of the Pseudomonas aeruginosa PAO1 Type IV pilus
Finally out in PLOS Pathogens. Congrats Hannah Ochner and co-authors. @mrclmb.bsky.social
journals.plos.org/plospathogen...

This Project Protein Runway sets proteins in the spotlight. The code is available through github.com/AndrewRadev/... . Publication will follow soon. #OpenScience #Bioinformatics #ProteinStructures #MolecularDynamics rant over and out. (n/n)
13.12.2024 14:38 — 👍 3 🔁 0 💬 0 📌 0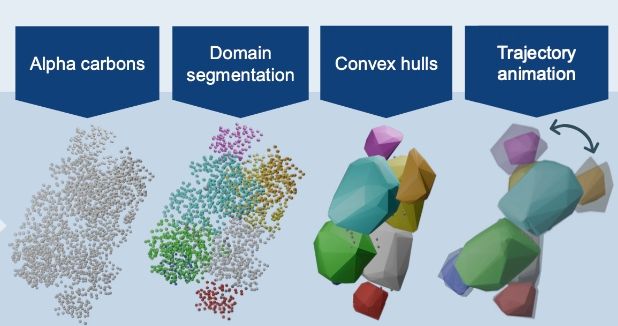
In the next phase additional domain segmentation methods will be implemented. In addition will use this to simulate and understand protein movements in more detail. (3/n)
13.12.2024 14:33 — 👍 1 🔁 0 💬 1 📌 0This phase is about visualization of protein dynamics, where proteins are first segmented into shapes followed by showing normal modes of the trajectories with help of Blender. Different segmentation algorithms were implemented in an OOP framework, to ensure extendability (2/n)
13.12.2024 14:17 — 👍 0 🔁 0 💬 1 📌 0Small rant. Funding rejected again. Quoting the reviewers "very ambitiius and original", "innovative" but "concerns .. about expertise". This semester, I started on the initial phase of this project nevertheless with a group of Master students and superhappy with results so far! (1/n)
13.12.2024 14:14 — 👍 4 🔁 0 💬 1 📌 0In our ab initio modeling group at imec we have two new PhD positions open
www.imec-int.com/en/work-at-i...
www.imec-int.com/en/work-at-i...
application deadline is December 15th.