So
```
pixi add r-gdalraster
```
is all you need to get going.
30.07.2025 07:28 — 👍 0 🔁 0 💬 0 📌 0
Linux containers != packaging. bsky.app/profile/matt...
Also, correct, you should have zero patience for that dev cycle, though until maybe 3 years ago this was realistically a way to do it for complex dependencies (e.g. CUDA). Now CUDA environments can be trivially handled with Pixi + conda-forge.
23.07.2025 13:34 — 👍 2 🔁 0 💬 1 📌 0
Strongly disagree with this. I've become far more of an expert in Linux containers in the last 7 years then I ever expected, and trying to compose environments between multiple images is not a viable solution for most people. Also, Linux containers != packaging! They're _distribution_ models.
23.07.2025 13:27 — 👍 2 🔁 0 💬 1 📌 2
You are correct that the feedstock creation for R on @conda-forge.org _needs_ to improve quite a bit (github.com/bgruening/co...). And once again with feeling, conda-forge is NOT Anaconda.com. There's no need to be using the defaults channel and can be trivially avoided.
23.07.2025 13:25 — 👍 1 🔁 0 💬 1 📌 0
System level binaries is an immediate non-starter. That is not how you build multiple computational environments.
23.07.2025 13:21 — 👍 0 🔁 0 💬 1 📌 0
Seems from this subthread that there's a potential for fixes in the future wrt PPM: bsky.app/profile/lach...
23.07.2025 05:56 — 👍 1 🔁 0 💬 1 📌 0
And for all the ones that don't? Which is most.
23.07.2025 05:30 — 👍 0 🔁 0 💬 1 📌 0
Without some big banner noting that there's additional required steps, those docs are wrong.
23.07.2025 05:29 — 👍 0 🔁 0 💬 0 📌 0
Posit Package Manager
If that's the case, that's good to hear. But I'd gently suggest that Posit fix the docs to note that Linux binary support is _not_ as described in the docs that you hit when you land on the PPM setup page.
packagemanager.posit.co/client/#/rep...
23.07.2025 05:29 — 👍 0 🔁 0 💬 1 📌 1
And so users should be able to get binaries for everything that Posit has built in PPM without having to run additional configuration scripts in advance, correct?
23.07.2025 05:19 — 👍 0 🔁 0 💬 2 📌 0
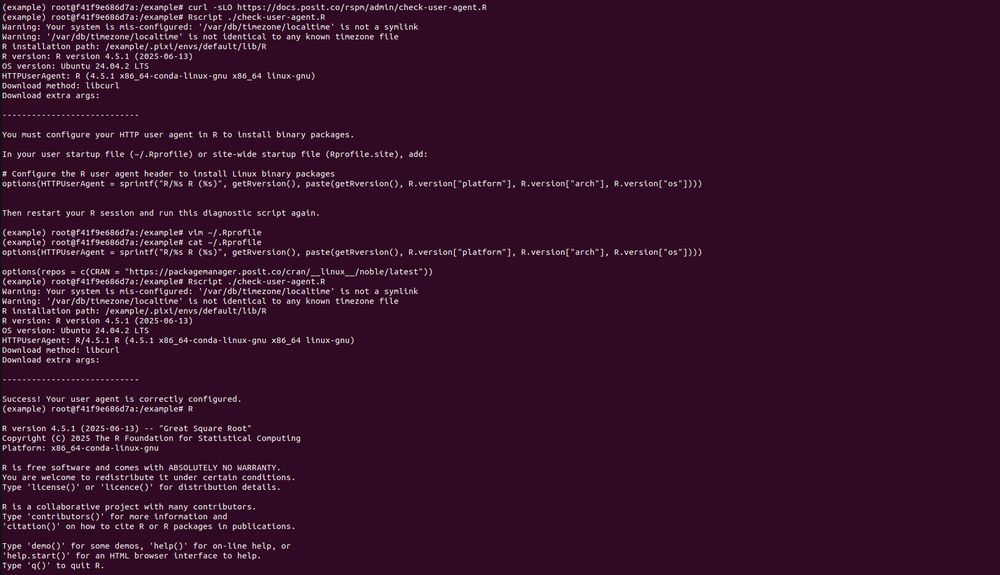
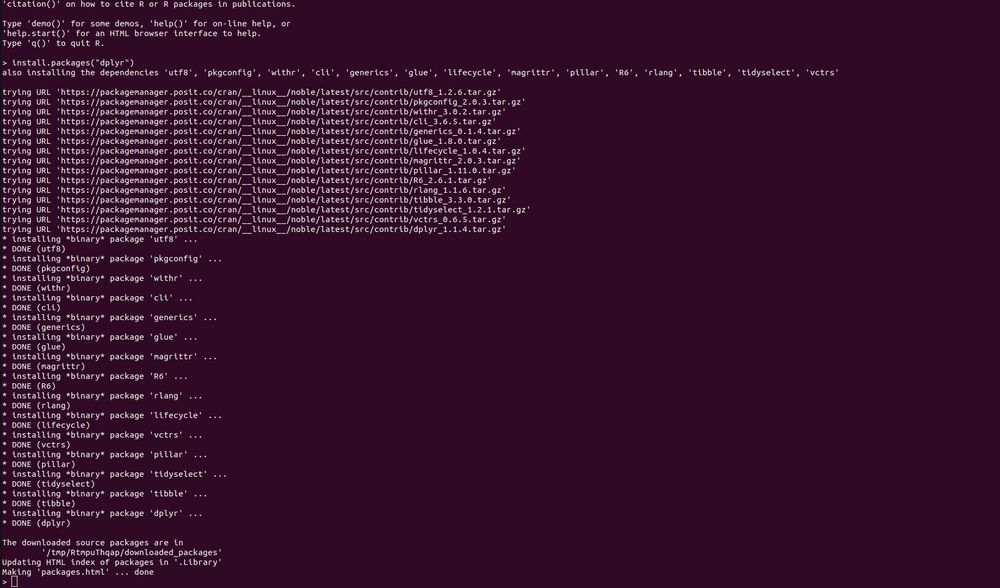
Thanks for the detailed summary, as that combination of things works. So if I understand things correctly, this is the required way to do things _now_, but whenever the next release of Package Manager happens there will be new docs on how to just provide an index URL in ~/.Rprofile to get binaries?
23.07.2025 05:19 — 👍 0 🔁 0 💬 1 📌 0
Again, real questions: Do R users primarily only use laptops then to do their work? Are people not running on clusters ever?
23.07.2025 04:34 — 👍 0 🔁 0 💬 1 📌 0
https://r-universe.dev/
Welcome to R-universe!
Aren't r-universe.dev indexes mutable? Doesn't that defeat any notion of reproducible environments? Or in practice is every build never removed?
23.07.2025 04:32 — 👍 0 🔁 0 💬 1 📌 0
Add a new CRAN-like repository — repo_add
Add a new repository to the list of repositories that pak uses to
look for packages.
The docs pak.r-lib.org/reference/re... make no mention of what the default behavior is. How should users know they need to use register a new repo? Is this your default workflow to always need to ask for "PPM@latest"?
23.07.2025 04:27 — 👍 1 🔁 0 💬 0 📌 0
* Correction. Ubuntu 24.04, though hopefully that doesn't matter at all.
23.07.2025 03:14 — 👍 0 🔁 0 💬 0 📌 0
Thanks very much for engaging and trying to point to examples of tools that you enjoy and help you. However, pak (or more posit package manager) fails to be able to provide binaries and so is not a solution: bsky.app/profile/matt...
23.07.2025 03:09 — 👍 0 🔁 0 💬 1 📌 0
Other important piece that I failed to mention: conda-forge was created in 2015 and started getting what I would call good around 2000, and then got _very_ good around 2022.
23.07.2025 03:06 — 👍 0 🔁 0 💬 0 📌 0
I'll comment here as that could be taken the wrong way: People obviously do real science on macOS and Windows! But scientific computing clusters are Linux, so R users doing computing at research institutions are either going to have to or _want_ to use Linux machines at some point.
23.07.2025 03:03 — 👍 0 🔁 0 💬 0 📌 0
I would be delighted for someone to show me that I've been missing a super obvious solution that everyone else knows, but from earlier responses to my original post months ago that doesn't seem to be the case. Thank you for taking so much time to engage and to point to things you use!
23.07.2025 03:02 — 👍 1 🔁 0 💬 1 📌 0
I am truly very happy to hear that for so many people on macOS and Windows the binary support for R packages on CRAN is excellent. That's great and should be celebrated! The question to me is "What do people doing serious computing with R on Linux (e.g. clusters) do for packages?"
23.07.2025 03:02 — 👍 1 🔁 0 💬 1 📌 0
RE: your past experience with conda: 100%. As a C++ & Python person I remember playing with conda in 2014 and rejecting it as the wheel support of Python packages started getting very good around the same time. I used to describe myself as a conda hater until about 2 years ago when I found Pixi!
23.07.2025 03:02 — 👍 1 🔁 0 💬 2 📌 0
My question then loops back to the original post I had. Why are people content with having basically no binary support for Linux?
23.07.2025 03:02 — 👍 1 🔁 0 💬 2 📌 0
> users generally don’t use the terminal much, so the idea of a separate package manager application seemed confusing.
That's super valid! I fully appreciate that if installing packages _inside_ of your code is the norm, then _external_ installation is very strange.
23.07.2025 03:02 — 👍 1 🔁 0 💬 1 📌 0
> What is the workflow you would like to see in R?
I'm not a real R user, so my views here aren't really important, but in general I'd like for people to be able to declaratively state what they need and have that and all dependencies get installed as securely built binaries very fast.
23.07.2025 03:02 — 👍 2 🔁 0 💬 1 📌 0
> I spend 5-10 minutes with pak to install dependencies, that feels fine.
I commend you for your patience. I am expecting full > 1GB environments of potentially complex dependencies to be solved, locked, downloaded, and installed in well under 1 minute.
23.07.2025 03:02 — 👍 1 🔁 0 💬 1 📌 0
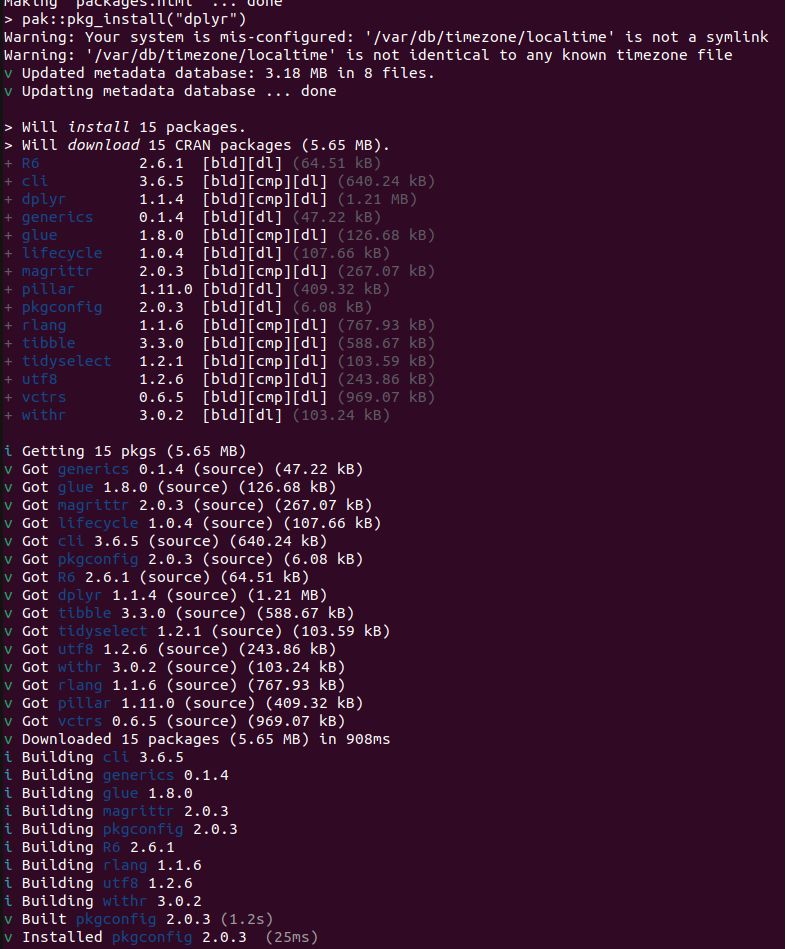
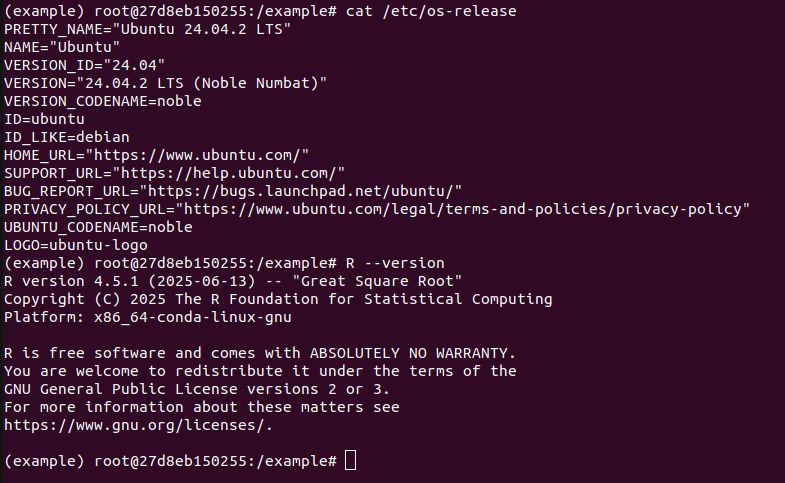
* What does
```
pak::pkg_install("dplyr")
```
do? It builds 15 packages from source. This is for a Ubuntu 22.04 x86 Linux container with R v4.5.x. So pak is not getting built distributions, because they don't exist. This is not a solution.
23.07.2025 03:02 — 👍 0 🔁 0 💬 3 📌 1
R Pak | Anaconda.org
* pak is on conda-forge (anaconda.org/conda-forge/...) but I'm going to try to use it how you are suggesting, which is off to a bad start because there's no binaries for it for Linux and required building from source 17 other packages. This is fine for me. Imagine you're a first year Ph.D. student.
23.07.2025 03:02 — 👍 1 🔁 0 💬 1 📌 0
To answer your questions across multiple posts:
* conda-forge gives you multi-platform built distributions of packages that can optionally have microarch optimizations. Having immutable built distributions is super important for multiple reasons, including scientific reproducibility.
23.07.2025 03:02 — 👍 1 🔁 0 💬 2 📌 0
Postdoc in Cvejic Group, BRIC, Københavns Universitet. PhD from Mayr Lab at Sloan Kettering Institute. Reproducible research enthusiast. Conda Forge R Team. Creator of scUTRquant.
|| PhD Statistics | Research Software Engineer | Loves #rstats | Outdoors person | Coffee person | Photography person | Serial Hobbiest | he/him ||
Builder. Collaborator. Open science & product nerd. Let’s make better tools for science communication.
Associate Professor of Earth, Environmental, and Planetary Science at Washington University in St. Louis • Planetary Data System Geosciences Node Director • Planetary Bastard • he/him/Sir
Incoherent superposition of education, writing, editing, consulting, outreach, and partnerships. Resonances include #author #teacher #LEGO #Physics #books www.youtube.com/drbenstill
Perpetually curious.
Scientist by training and temperament.
Tennis player. Ocean lover. Occasional mischief-maker.
Views mine.
#Duke #UCSD Alum; Recovering Academic; Research IT
@jacksonlab.bsky.social; core dev @napari.fosstodon.org.ap.brid.gy
they/them — building Python developer tools in Rust at Astral
Enthusiastic dev, Python/C++/Rust
Core member of conda-forge
Maintainer of grayskull
computational physicist • accelerates accelerator research at @berkeleylab.bsky.social • laser-plasma aficionado • PByte connoisseur • mostly memory BW bound • openPMD • sciency C++ & 🐍 • GPGPU since before it was cool • he/his • Dr. rer. nat. Dipl.-Phys.
Front End Instructor at Madison College
Neutral Good
Formerly: Ionic, Teal Media, Spyder Trap
Data Scientists Professional working at the Data Science Institute UW Madison🔬
Stats/ML/Math eg Topology and Dynamical Systems
Team player!
https://mariaorosdatascientist.netlify.app/
author of THE MILITIA HOUSE (Henry Holt, 23) | Sewanee Writers’ Conference fellow | Shirley Jackson Award nominee | Center for Fiction longlist | Illinois Bois ➡️ Florida Man | https://www.johnmilas.com/
Research Scholar at Yale Law School studying the history of science in China and US-China relations. Particle physicist by training. Writer at various places. Editor at Made in China Journal. Co-host of Dissident at the Doorstep.
Chancellor's Distinguished Professor of Physics, UCSD. Interested in everything.
Theoretical physicist researching condensed & living matter, fluid mechanics, statistical mechanics. Bad at staying in my lane.
https://guava.physics.ucsd.edu/~nigel/
Director, Open Science Strategy @ HHMI
Professor a NYU; Chief AI Scientist at Meta.
Researcher in AI, Machine Learning, Robotics, etc.
ACM Turing Award Laureate.
http://yann.lecun.com
Science Philanthropy, Open Science, and Photography. Leave me anonymous feedback! → https://www.admonymous.co/karthik
A team of open-source maintainers and engineers providing solutions and support for open-source projects used by millions worldwide. #Jupyter, #condaforge, #Mamba, #ApacheArrow, #Xtensor, and more.
 30.07.2025 07:51 — 👍 0 🔁 0 💬 0 📌 0
30.07.2025 07:51 — 👍 0 🔁 0 💬 0 📌 0