I wrote a new blog post on using the logit lens to interpret protein language models!
liambai.com/logit-lens/
It has some interactive visualization you can play around with.
22.05.2025 20:08 — 👍 0 🔁 0 💬 0 📌 0
It's long seemed that molecular biology is a natural home for ML interpretability research, given the maturity of human-constructed models of biological mechanisms—permitting direct comparison with their ML-derived counterparts—unlike vision and NLP. Our first foray below👇.
10.02.2025 16:15 — 👍 38 🔁 15 💬 0 📌 0
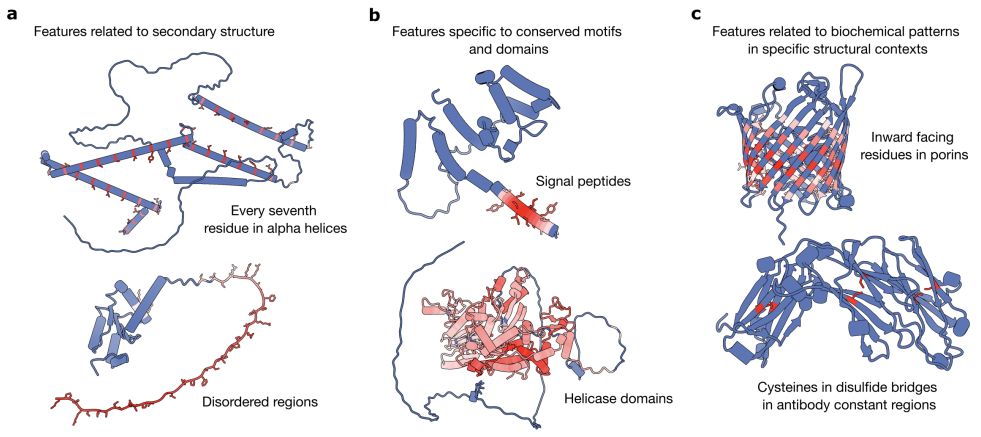
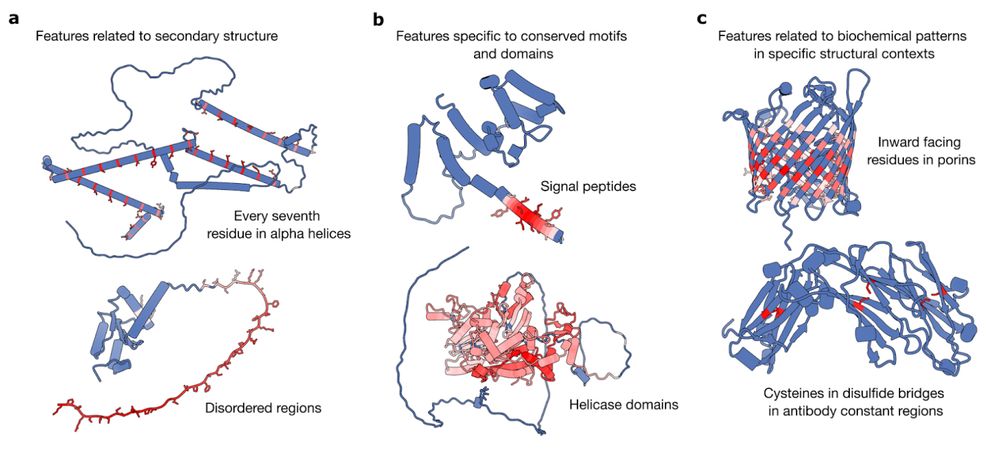
A range of features identified from sparse autoencoders trained on different layers on ESM2-650M
This might be the best paper on applying sparse autoencoders to protein language models. The authors identify how neural networks trained on amino acid sequences "discover" different features, some specific to individual protein families, other for substructures
www.biorxiv.org/content/10.1...
10.02.2025 12:15 — 👍 44 🔁 11 💬 2 📌 0
Can we learn protein biology from a language model?
In new work led by @liambai.bsky.social and me, we explore how sparse autoencoders can help us understand biology—going from mechanistic interpretability to mechanistic biology.
10.02.2025 16:12 — 👍 44 🔁 24 💬 2 📌 1
Discover the Languages of Biology
Build computational models to (help) solve biology? Join us! https://www.deboramarkslab.com
DM or mail me!
Professor, Programmer in NYC.
Cornell, Hugging Face 🤗
med chem @ Arvinas | PhD @ Columbia | hiker | cocktail enthusiast | he/him/huz/dad | dreaming of Sedona | while I have a connection to Arvinas, all opinions expressed are my own and do not represent the views, opinions, or positions of the company
AI for Life Sciences & Healthcare at NVIDIA | trained as a scientist from JGI, UW & UC Berkeley | views are all mine
Probabilistic machine learning to address questions in evolution and health #EvolutionaryMedicine. PI at the Centre for Genomic Regulation, co-leading a group with Mafalda Dias. Previously Harvard.
Genomics, Machine Learning, Statistics, Big Data and Football (Soccer, GGMU)
DeepMind Professor of AI @Oxford
Scientific Director @Aithyra
Chief Scientist @VantAI
ML Lead @ProjectCETI
geometric deep learning, graph neural networks, generative models, molecular design, proteins, bio AI, 🐎 🎶
Scientist, Assistant Professor at MIT biology, #FirstGen
AI4Science researcher. Associate Professor @CSHL. My lab advances AI for genomics and healthcare!
http://koo-lab.github.io
PhD student @MIT • Research on Generative Models for Biophysics and Drug Discovery
MIT PhD Student - ML for biomolecules - https://hannes-stark.com/
Machine learning for biology | Stanford and Arc Institute
Principal Researcher in BioML at Microsoft Research. He/him/他. 🇹🇼 yangkky.github.io
Bren Professor of Computational Biology @Caltech.edu. Blog at http://liorpachter.wordpress.com. Posts represent my views, not my employer's. #methodsmatter
Developing data intensive computational methods • PI @ Seoul National University 🇰🇷 • #FirstGen • he/him • Hauptschüler
Designing peptides/proteins to program biology! 🧬💻🧫 Assistant Professor at Duke | Co-Founder of Gameto and UbiquiTx | MIT SB, SM, PhD
Google Chief Scientist, Gemini Lead. Opinions stated here are my own, not those of Google. Gemini, TensorFlow, MapReduce, Bigtable, Spanner, ML things, ...
Computational biology fellow w Jose Maria Carazo and Carlos Oscar Sorzano at CNB, Madrid, working on protein dynamics and cryoem (done Marie Sklododowska-Curie, now CSIC grant). Did postdoc w Ivet Bahar. Neuro and covid
Protein engineering & synthetic biochemistry at GSK
Opinions my own
https://linktr.ee/ddelalamo
MLing biomolecules en route to structural systems biology. Asst Prof of Systems Biology @Columbia. Prev. @Harvard SysBio; @Stanford Genetics, Stats.

