
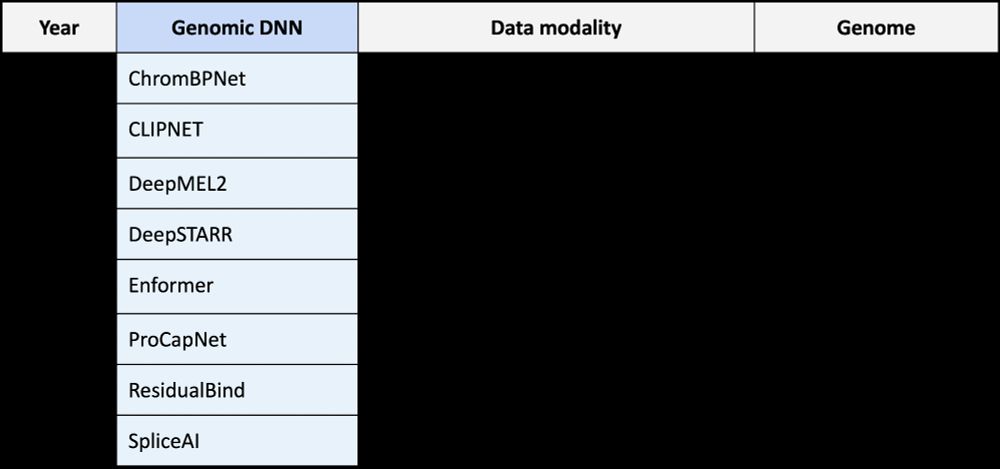
90th Symposium: AI in Biology
Cold Spring Harbor Laboratory Meetings & Courses -- a private, non-profit institution with research programs in cancer, neuroscience, plant biology, genomics, bioinformatics.
Save the date: The premier AI x Bio meeting of 2026 will be held at CSHL on May 26-31!
The program brings together 50+ invited leaders in genomics, transcriptomics, protein design, drug discovery, neuro-AI, pathology, agentic AI, and more!
Abstract due: March 26
meetings.cshl.edu/meetings.asp...
04.01.2026 00:03 —
👍 10
🔁 0
💬 0
📌 0

Beware of LLM blindspots. #AI4Science
08.11.2025 21:24 —
👍 5
🔁 1
💬 0
📌 0


Yijie Kang (CSHL, Stony Brook) from @pkoo562.bsky.social Lab presented "Decoding the sequence basis of Pol II elongation with deep learning"
07.11.2025 15:05 —
👍 2
🔁 1
💬 1
📌 0

Exciting symposium on AI and Biology at EMBO | EMBL in Heidelberg on 10-13 March 2026!
Excellent lineup of invited speakers across various scales of biology!
Deadline for abstract submission is coming up — Dec 2.
🔗 www.embl.org/about/info/c...
#EESAIBio @EMBLEvents
07.11.2025 00:16 —
👍 9
🔁 7
💬 0
📌 1
We find regulatory DNA is readily reprogrammable with a few key mutations! We observed similar phenomenon across all genomic DNNs we tested! 12/N
09.10.2025 12:08 —
👍 0
🔁 0
💬 1
📌 0
SEAM is a powerful tool that helps to: 1) explore the high evolvability landscape of regulatory sequences; 2) identifies mutations that drive mechanistic changes; and 3) dissect motif syntax and context dependencies. 11/N
09.10.2025 12:08 —
👍 0
🔁 0
💬 1
📌 0

And we tested the backgrounds across different ChromBPNet models independently trained on DNase-seq and ATAC-seq and we observe similar backgrounds! This suggests these mutagenesis-robust patterns are important context that reflects properties of the local sequence space. 10/N
09.10.2025 12:08 —
👍 0
🔁 0
💬 1
📌 0
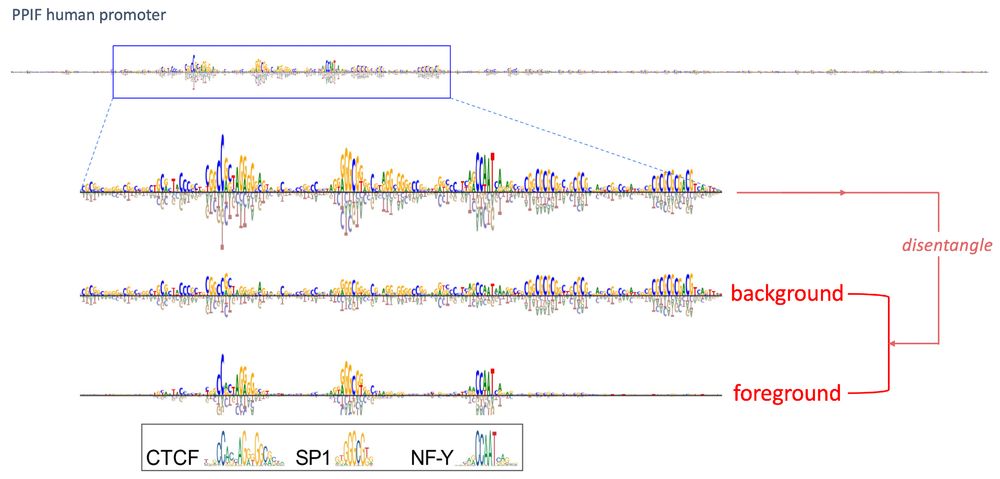
While previous analyses focused on differences in attr maps across clusters, a surprising observation was that there were also shared patterns. We disentangled the attribution signals that are sensitive versus robust to mutagenesis – we call them foreground and background. 9/N
09.10.2025 12:08 —
👍 0
🔁 0
💬 1
📌 0

This analysis flagged 2 key mutations at positions 170 & 174 that created a new CAAT box. To test necessity & sufficiency, we mutated each individually and together, then examined attr maps+ predictions
- single mutations -> no change
- double mutation -> CAAT box + new Inr
8/N
09.10.2025 12:08 —
👍 0
🔁 0
💬 1
📌 0

Applying SEAM to CLIPNET, which predicts transcriptional activity measured via PRO-cap, we find that many SNVs lead to new clusters in the PIK3R3 promoter. A few specific mutations can quantitatively tune gene expression and SEAM can find them! 7/N
09.10.2025 12:08 —
👍 1
🔁 0
💬 1
📌 0
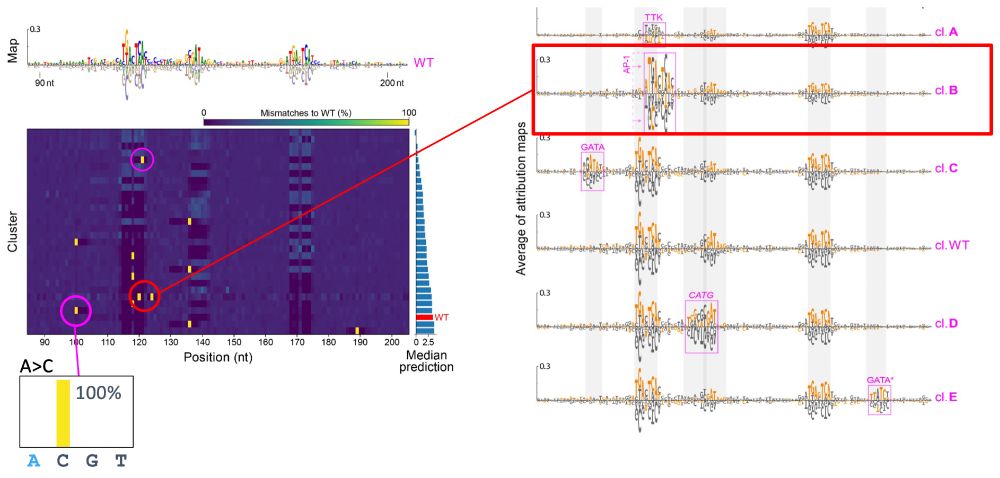
Now, if we plot the percent mismatch of the nucleotides with respect to WT for each cluster, you can see yellow bars that reflect all sequences in the cluster share the same single nucleotide mutation. This analysis pinpoints the exact mutation that led to the new mechanism! 6/N
09.10.2025 12:08 —
👍 0
🔁 0
💬 1
📌 0

Low entropy reflects all the sequences share the same nucleotides, while high entropy reflects different mutations destroyed the motif. Sometimes, we see motif preserving signature outside the vertical bands. This represents a de novo motif that appeared within that cluster. 5/N
09.10.2025 12:08 —
👍 1
🔁 0
💬 1
📌 0


If we calculate the positional entropy of the sequences within each cluster, we get a cluster summary matrix. The vertical bands highlight the locations of the motifs in WT seq and entropy levels indicate whether the motif is present or not in the attr maps in each cluster. 4/N
09.10.2025 12:08 —
👍 1
🔁 0
💬 1
📌 0
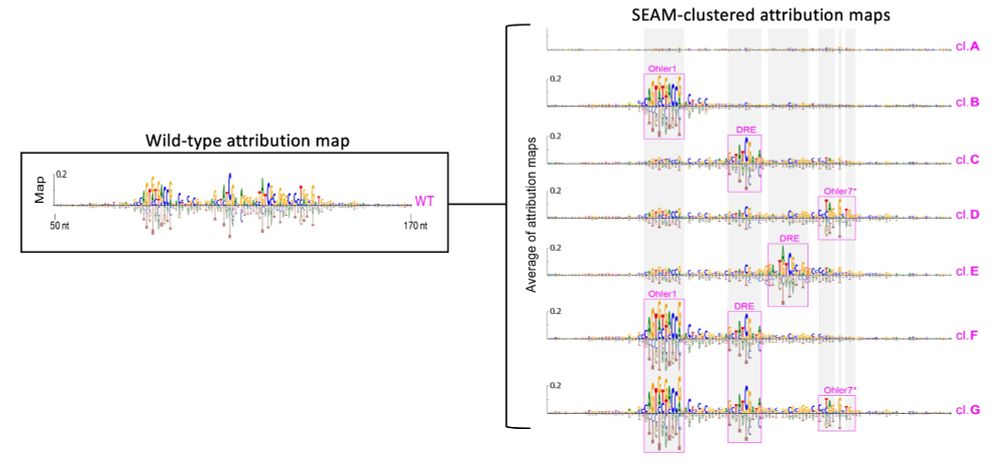
Attr maps can sometimes be easy to interpret, and sometimes they're complex. SEAM's clustered attr maps are cleaner (think SmoothGrad) and they decompose complex mechanisms via partial random mutagenesis, which occasionally disrupts key binding sites. 3/N
09.10.2025 12:08 —
👍 1
🔁 0
💬 1
📌 0

SEAM is conceptually simple. Starting from a reference sequence:
1) sample in a local region of sequence space via partial random mutagenesis
2) calculate attr maps to unveil the mechanisms
3) cluster attr maps based on shared mechanisms
4) cluster-based sequence analysis
2/N
09.10.2025 12:08 —
👍 1
🔁 0
💬 1
📌 0

Which mutations rewire function of regulatory DNA?
Excited to share SEAM: Systematic Explanation of Attribtuion-based Mechanisms. SEAM is an explainable AI method that dissects cis-regulatory mechanisms learned by seq2fun genomic deep learning models.
Led by @EESetiz
1/N 🧵👇
09.10.2025 12:02 —
👍 29
🔁 10
💬 1
📌 0

Congratulations to John Clarke, Michel Devoret and John Martinis on receiving the 2025 Nobel Prize in Physics!
www.nobelprize.org/prizes/physi...
I have fond memories of my time in the Clarke lab, where I did my Honors Thesis on ultra low-field MRI w/ SQUIDs as an undergrad at UC Berkeley!
07.10.2025 14:16 —
👍 9
🔁 1
💬 0
📌 0
Check out a Research Highlights on our work at @naturemethods by Lin Tang!
www.nature.com/articles/s41...
19.09.2025 16:36 —
👍 7
🔁 0
💬 0
📌 0

Richard Bonneau giving the last keynote on navigating the complexity of drug discovery and their lab-in-the-loop for molecule design! #MLCB
11.09.2025 17:40 —
👍 3
🔁 0
💬 0
📌 0

First talk a (surprise) keynote by Jacob Schreiber from UMass Medical talking about fruit-themed AI tools for understanding and designing regulatory DNA
11.09.2025 13:44 —
👍 3
🔁 0
💬 0
📌 0
2025 MLCB day 2 is starting now!
Streaming live now!
m.youtube.com/watch?v=PxlXNb…
11.09.2025 13:42 —
👍 2
🔁 0
💬 1
📌 0

Now Barbara Engelhardt giving a keynote on characterizing behaviors of modified T cells in live cell imaging data using machine learning!
10.09.2025 17:58 —
👍 4
🔁 0
💬 0
📌 0

Next talk by Courtney Shearer who is talking about genomic language models for zero shot promoter indel effects!
10.09.2025 15:15 —
👍 1
🔁 0
💬 1
📌 0

Next talk by Alan Murphy and Masayuki (Moon) Nagai (from my lab!) who are talking about how naive fine-tuning genomic DNNs leads to catastrophic forgetting and propose *iterative causal refinement* to improve learned associations to causal understanding of cis-regulatory biology!
10.09.2025 14:53 —
👍 0
🔁 0
💬 1
📌 0

Next talk by Johannes Linder at Calico. Talking about expanding genomic seq2fun DNNs with RBP binding and RNA processing data to consider post-transcriptional regulation.
10.09.2025 14:38 —
👍 1
🔁 0
💬 1
📌 0
Some technical delays but we are all set!
First talk by Alexis Battle! @alexisbattle.bsky.social
10.09.2025 13:52 —
👍 6
🔁 0
💬 1
📌 0


























