
Tomorrow we have a reading group session with @nscorley and @SimMat20 about "Accelerating Biomolecular Modeling with AtomWorks and RF3" www.biorxiv.org/content/10.1...
We will also discuss how to close the gap between AF3 and its open source replications!
portal.valencelabs.com/starklyspeak...
05.10.2025 20:21 — 👍 8 🔁 0 💬 0 📌 0
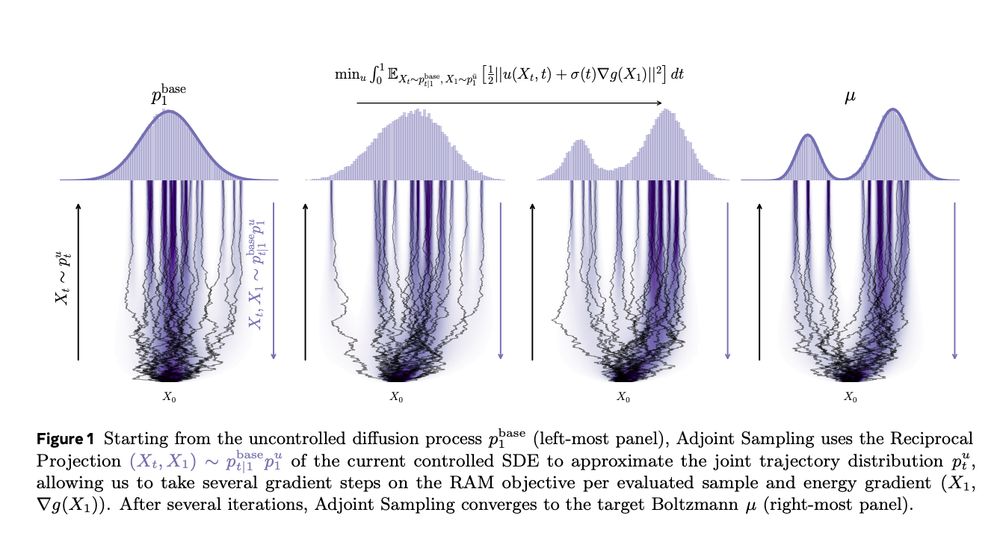
Tomorrow we discuss diffusion models for sampling unnormalized densities "Adjoint Sampling: Highly Scalable Diffusion Samplers via Adjoint Matching"
arxiv.org/abs/2504.11713
Join us on zoom at 9am PT / 12pm ET / 6pm CEST: portal.valencelabs.com/starklyspeak...
03.08.2025 23:49 — 👍 18 🔁 3 💬 0 📌 0
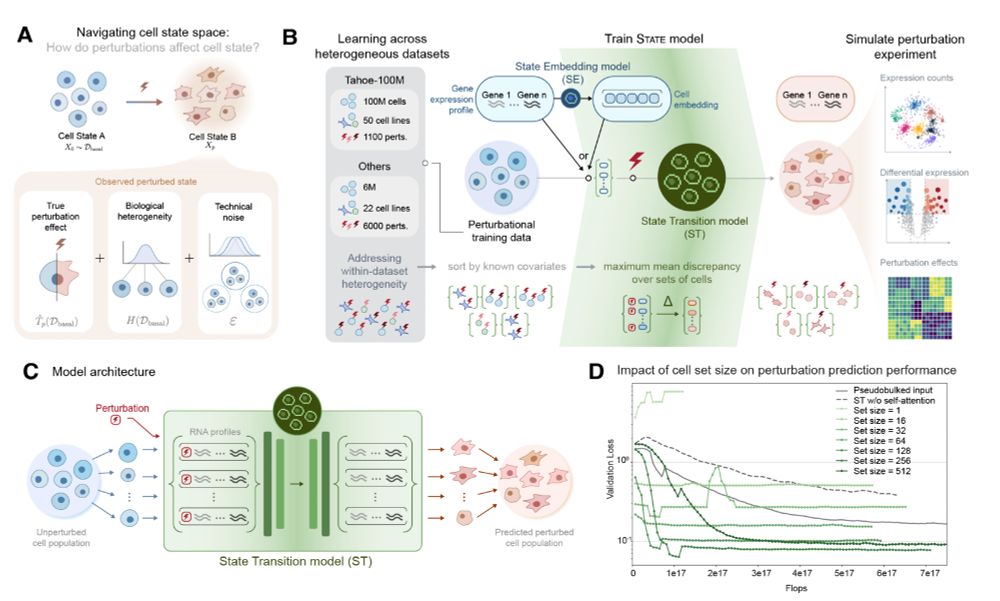
In 1h we discuss "Predicting cellular responses to perturbation across diverse contexts with State" in the reading group with the author Abhinav Adduri! www.biorxiv.org/content/10.1...
Join us on zoom at 9am PT / 12pm ET: portal.valencelabs.com/starklyspeak...
21.07.2025 15:02 — 👍 4 🔁 0 💬 0 📌 0
Starkly Speaking tomorrow: @bwood_m will present "UMA: A Family of Universal Models for Atoms" ai.meta.com/research/pub...
Join us on Zoom 12pm ET / 6pm CEST: portal.valencelabs.com/starklyspeak...
29.06.2025 19:52 — 👍 3 🔁 0 💬 0 📌 0

Monday Starkly Speaking: we understand a core diffusion model ingredient better - classifier free guidance.
Via "Classifier-Free Guidance: From High-Dimensional Analysis to Generalized Guidance Forms" arxiv.org/abs/2502.07849
On Zoom 12pm ET / 6pm CEST: portal.valencelabs.com/starklyspeak...
21.06.2025 23:42 — 👍 8 🔁 1 💬 0 📌 0
YouTube video by Valence Labs
Boltz-2: Towards Accurate and Efficient Binding Affinity Prediction
Presentation of Boltz-2 by @pas_saro @GabriCorso @jeremyWohlwend!
Structure models and careful affinity data handling result in an affinity predictor approaching FEP accuracy at 1000x the speed.
www.youtube.com/watch?v=iHDa...
18.06.2025 22:19 — 👍 4 🔁 1 💬 0 📌 0
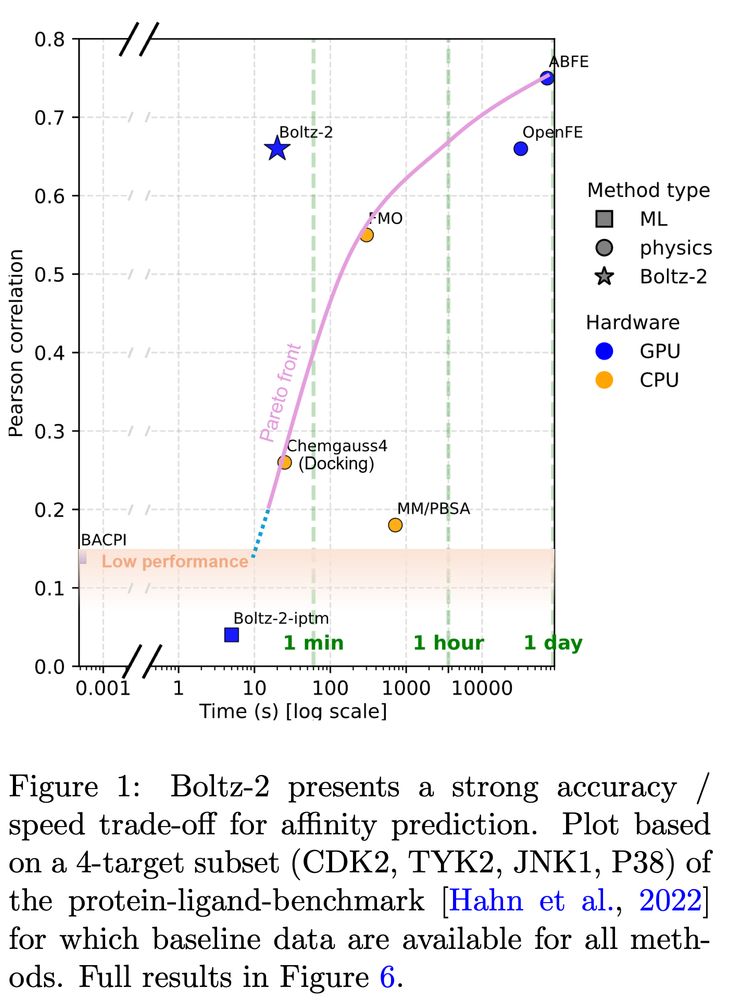
On Tue 12pm ET on Zoom: Saro Passaro, Gabriele Corso, Jeremy Wohlwend will present Boltz-2!
bit.ly/boltz2-pdf
Structure models and careful affinity data handling result in an affinity predictor approaching FEP accuracy at 1000x the speed.
Zoom link: portal.valencelabs.com/starklyspeak...
14.06.2025 17:14 — 👍 5 🔁 1 💬 0 📌 0
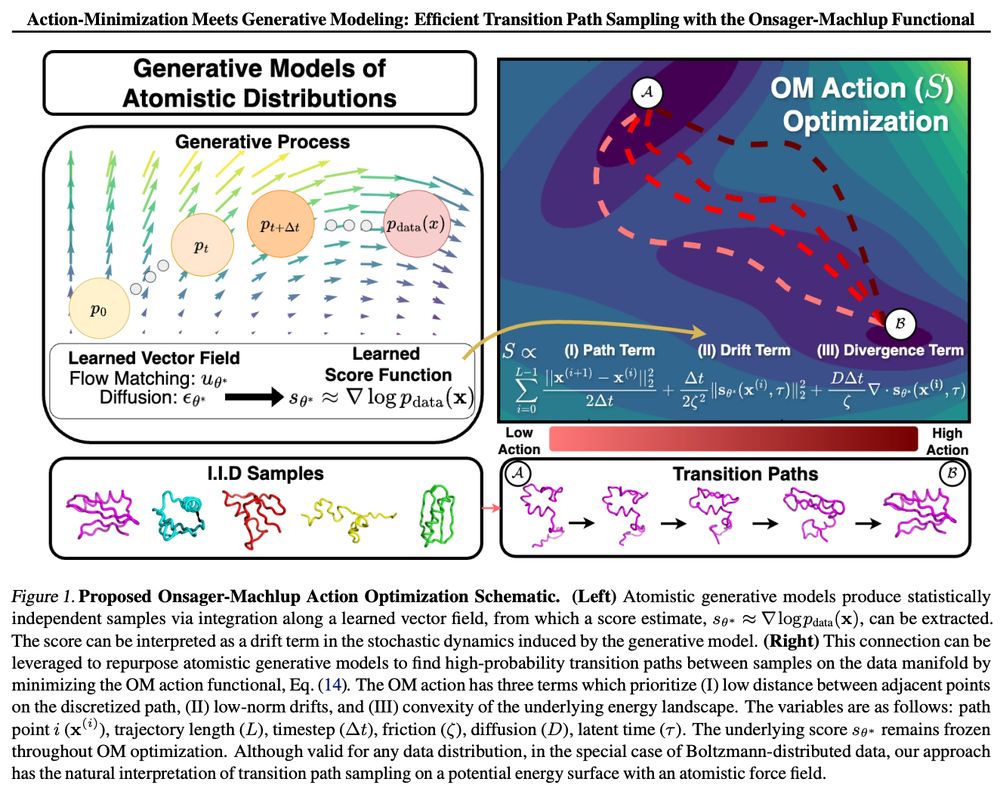
Tomorrow in our reading group we will discuss "Action-Minimization Meets Generative Modeling: Efficient Transition Path Sampling with the Onsager-Machlup Functional" arxiv.org/abs/2504.18506
On zoom 9am PT / 12pm ET / 6pm CEST: portal.valencelabs.com/starklyspeak...
08.06.2025 19:43 — 👍 10 🔁 0 💬 0 📌 0
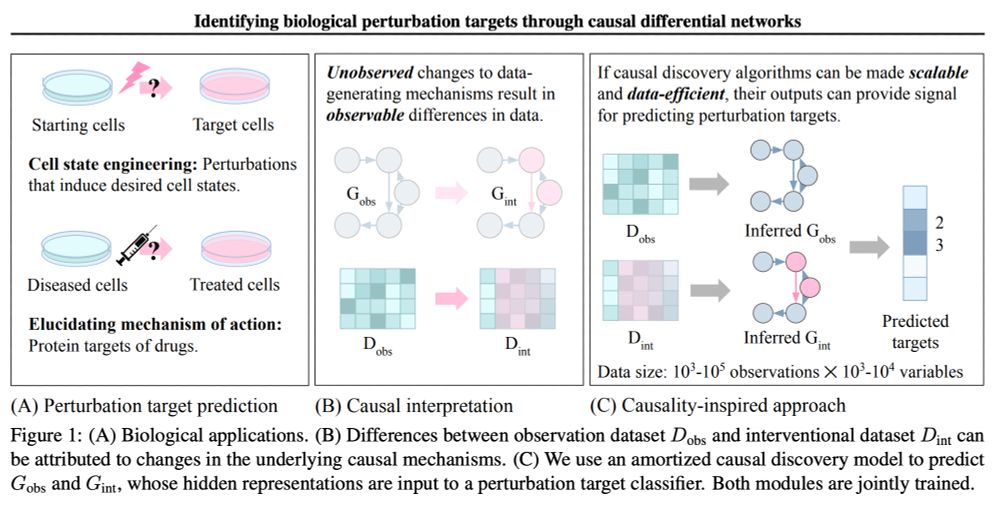
Reading group tomorrow: Identifying perturbation targets through causal differential networks arxiv.org/abs/2410.03380
With @menghua_wu!
Join us on Zoom at 9am PT 12pm ET 6pm CEST: portal.valencelabs.com/starklyspeak...
01.06.2025 20:58 — 👍 3 🔁 0 💬 0 📌 0

Graphic that reads "MoML @ MIT Call for Papers | 6th Molecular Machine Learning Conference" followed by smaller text that says "Topics of interest include but are not limited to: Geometric deep learning • Graph machine learning • Molecular dynamics • Structure prediction • Machine learning for quantum chemistry"At the bottom third of the graphic, the text reads "Deadline: Friday, Sept. 26, 2025 | Conference Date: Wednesday, Oct. 22, 2025 | Win a cash prize of $1,526 for Best Paper!"
📢 Thrilled to announce the return of MoML @ MIT on Oct. 22 and that paper submissions are now open! Students & postdocs w/ accepted papers will be granted FREE admission to attend and have the chance at winning the Octavian-Eugen Ganea Prize for Best Paper! 🧵 #AcademicSky #compchem #drugdesign
28.05.2025 20:28 — 👍 6 🔁 4 💬 1 📌 0
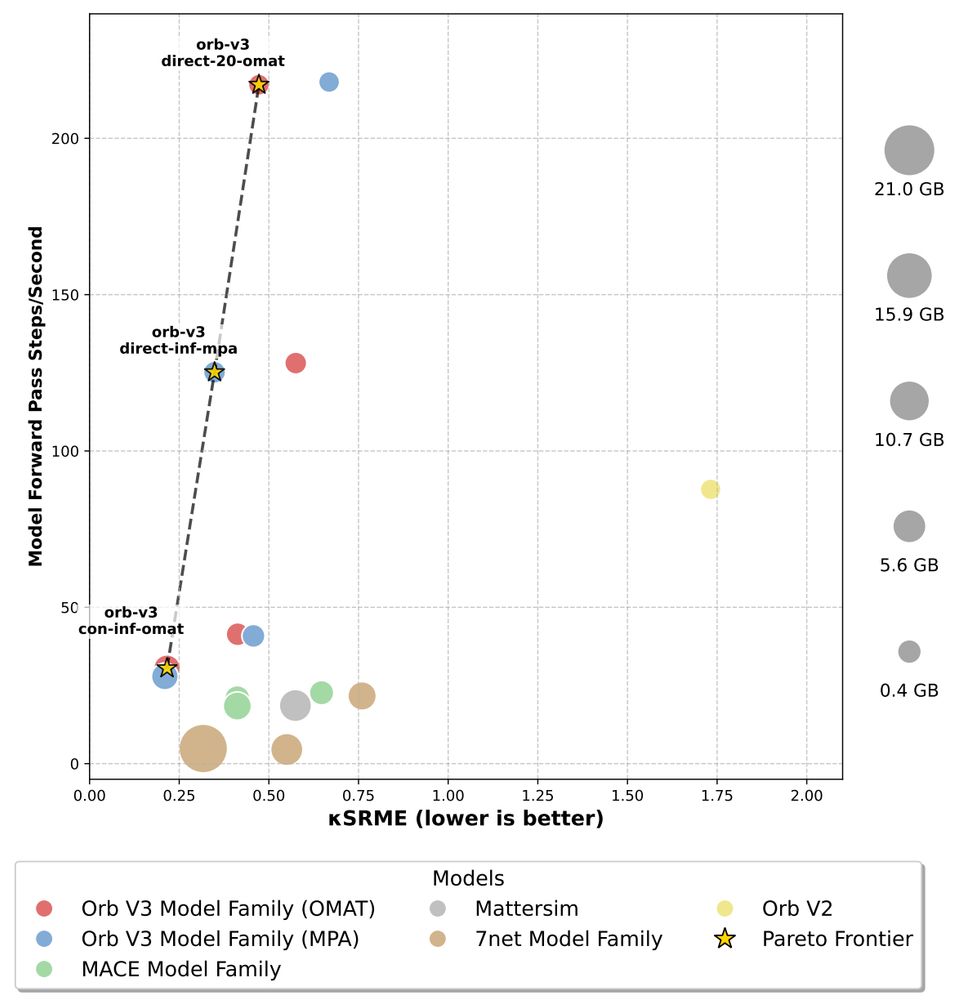
Tomorrow @TimothyDuignan joins us to discuss "Orb-v3: atomistic simulation at scale" arxiv.org/abs/2504.06231 and surrounding models to get our understanding of that field up to date!
On Zoom 9am PT / 12pm ET / 6pm CEST: portal.valencelabs.com/starklyspeak...
25.05.2025 18:12 — 👍 8 🔁 2 💬 0 📌 0
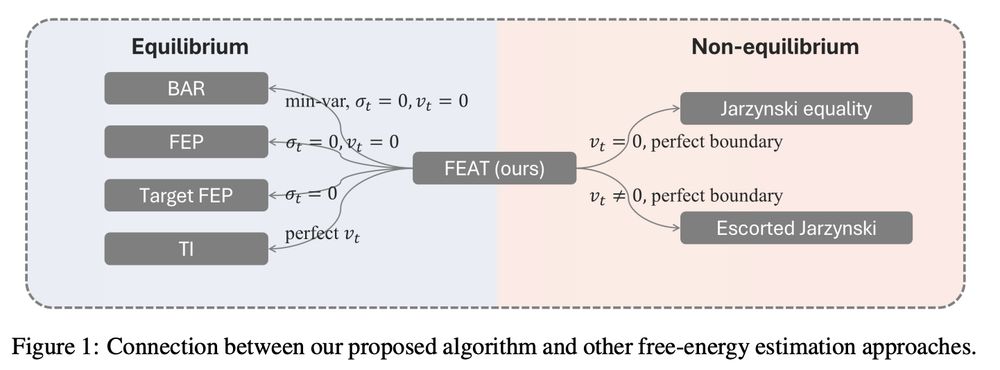
Tomorrow the authors @YuanqiD and Jiajun He will present their paper "FEAT: Free energy Estimators with Adaptive Transport" arxiv.org/abs/2504.11516
Estimating FED is a strong tool for comparing drug's binding affinities
Join us on zoom at 12pm ET / 6pm CEST: portal.valencelabs.com/starklyspeak...
18.05.2025 18:40 — 👍 5 🔁 0 💬 0 📌 0
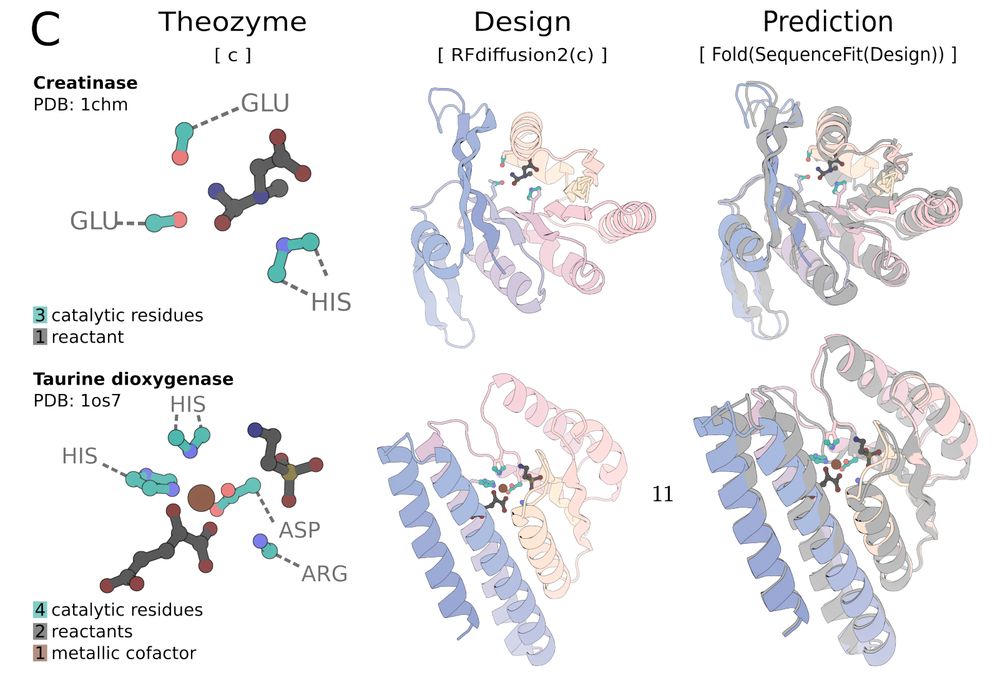
Reading group tomorrow: @json_yim and @woodyahern present "Atom level enzyme active site scaffolding using RFdiffusion2" www.biorxiv.org/content/10.1...
Join on Zoom at 9am PT / 12pm ET / 6pm CEST: portal.valencelabs.com/starklyspeak...
11.05.2025 17:11 — 👍 12 🔁 3 💬 0 📌 0
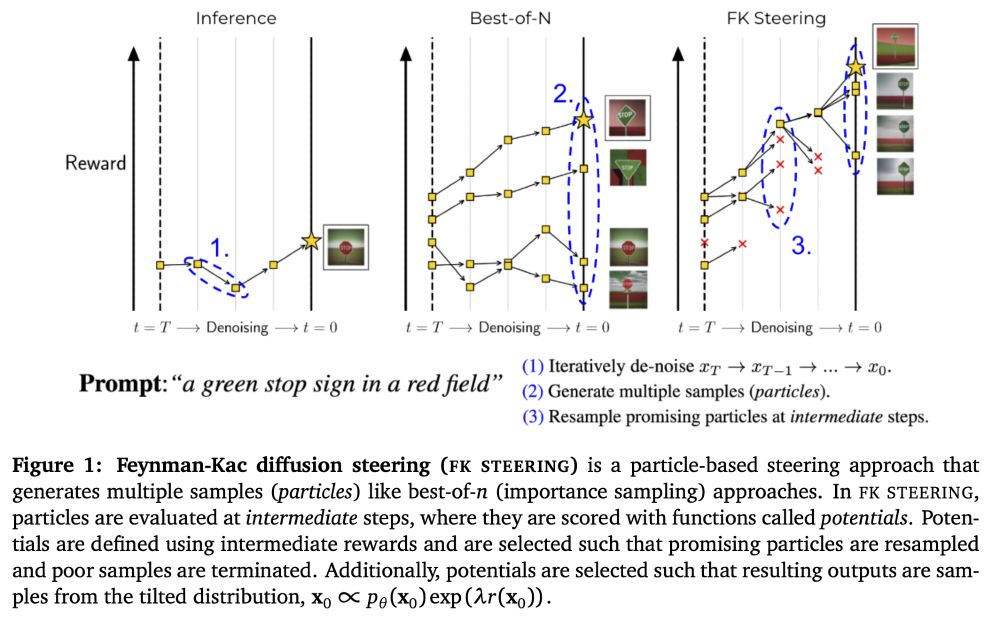
In the reading group sessions tomorrow Ezra Erives, I and whoever joins, will discuss "A General Framework for Inference-time Scaling and Steering of Diffusion Models" arxiv.org/abs/2501.06848
On zoom at 9am PT / 12pm ET / 6pm CET: portal.valencelabs.com/logg
20.04.2025 16:47 — 👍 14 🔁 2 💬 0 📌 0

Reading group session tomorrow, March 24th, is at 3pm PT / 6pm ET / midnight CEST about:
"One-step Diffusion Models with f-Divergence Distribution Matching" arxiv.org/abs/2502.15681
Presented by @xuyilun2
Zoom link: portal.valencelabs.com/logg
23.03.2025 21:47 — 👍 4 🔁 0 💬 0 📌 0
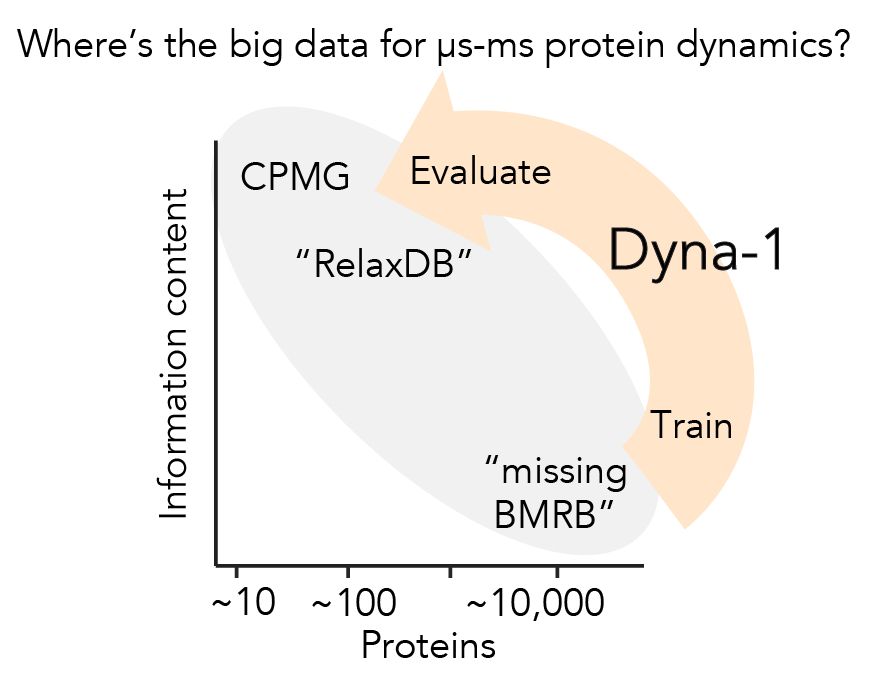
Protein dynamics was the first research to enchant me >10yrs ago, but I left in PhD bc I couldn't find big experimental data to evaluate models.
Today w @ginaelnesr.bsky.social, I'm thrilled to share the big dynamics data I've been dreaming of, and the mdl we trained w them: Dyna-1.
📝: rb.gy/de5axp
20.03.2025 15:02 — 👍 84 🔁 25 💬 2 📌 2
Check out this talk from our own Garyk Brixi talking about Evo 2!
14.03.2025 14:35 — 👍 8 🔁 4 💬 0 📌 0
YouTube video by Valence Labs
Genome modeling and design across all domains of life with Evo 2 | Garyk Brixi
Video is up!
"Genome modeling and design across all domains of life with Evo 2" youtu.be/Rarn97Wpl1A
With the author Garyk Brixi!
13.03.2025 23:55 — 👍 18 🔁 3 💬 0 📌 1
The code & camera-ready version of our #ICLR2025 paper on "Multi-domain Distribution Learning for De Novo Drug Design" are now available
📚 Paper: openreview.net/forum?id=g3V...
💻 Code: github.com/LPDI-EPFL/Dr...
(1/4)
07.03.2025 13:38 — 👍 25 🔁 7 💬 2 📌 1
🚨 Check out DrugFlow, our new generative model for structure-based drug design. DrugFlow provides an atom-level confidence score for each designed molecule, and can adjust molecular size on the fly!
Additional details in thread 🧵
#ICLR2025
11.03.2025 15:02 — 👍 23 🔁 7 💬 1 📌 0
🔥 ProtComposer (ICLR'25 Oral) is a Swiss Army knife:
(i) Manually create new protein structure layouts? ✅
(ii) Generation with favorable designability/diversity/novelty trade-offs? ✅
(iii) Spatially edit given proteins? ✅
Very original work by the amazing @hannes-stark.bsky.social and Bowen Jing!🔥
11.03.2025 01:07 — 👍 7 🔁 2 💬 0 📌 0

Code: github.com/NVlabs/protc...
Humans:
Moi*, Bowen Jing*, Tomas Geffner, @jyim.bsky.social , Tommi Jaakkola, Arash Vahdat, @karstenkreis.bsky.social
Many of us will be at #ICLR2025 with this stuff, happy to chat!
Also happy to improve the NVIDIA logo a bit with this beauty made by Bowen🙃
6/6
10.03.2025 19:51 — 👍 4 🔁 0 💬 0 📌 0
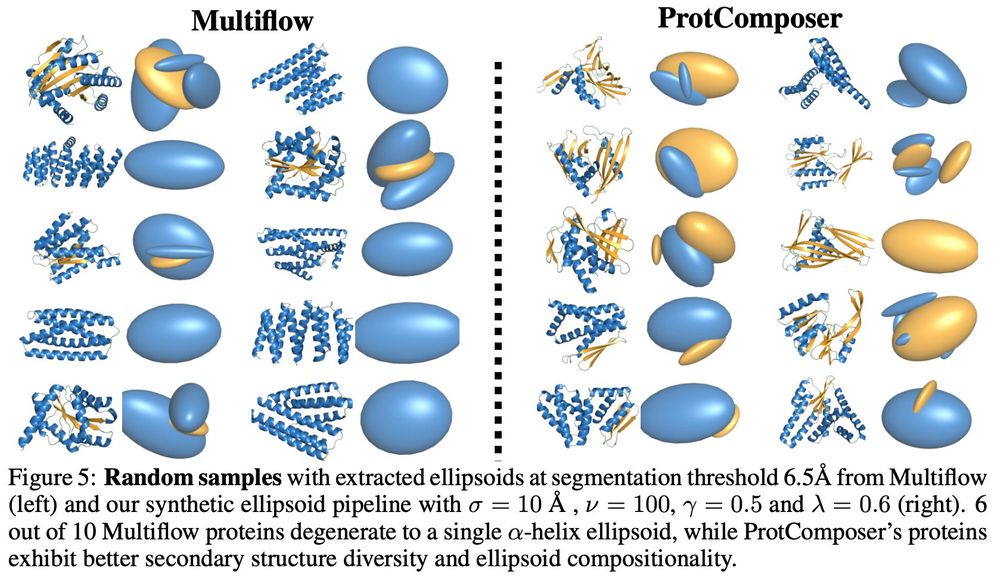
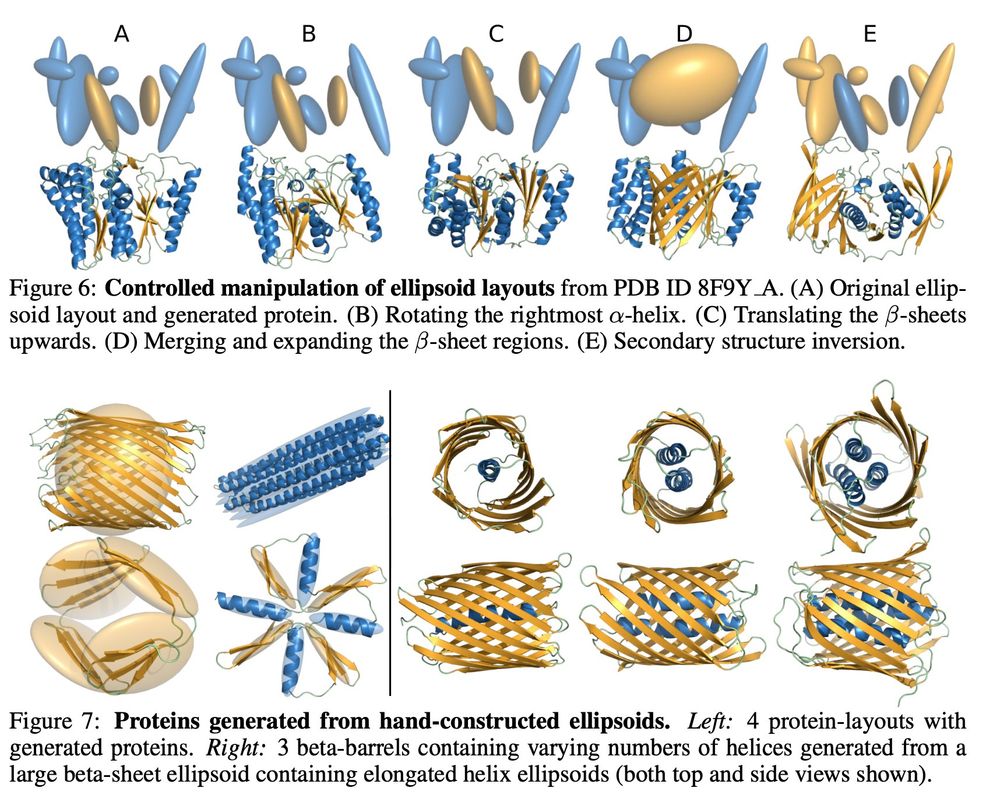
Here are images of the rest of the figures because why not.
(we also have classifier free guidance between the conditional and the unconditional model to trade off ellipsoid adherence or to recover the original model)
5/6
10.03.2025 19:51 — 👍 2 🔁 0 💬 1 📌 0
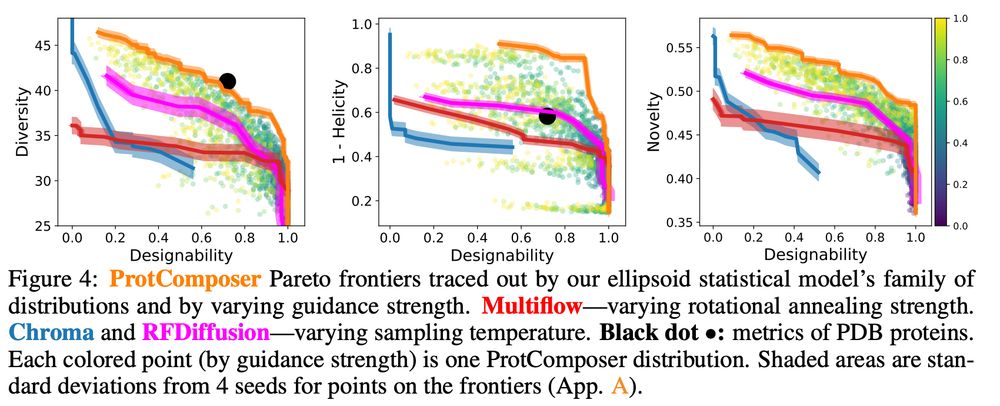
Our ellipsoid generation model is a simple statistical model => not overfit on the data => produces novel and diverse ellispoid layouts => ProtComposer produces novel and diverse proteins => nice pareto frontiers of designability vs. diverisity/novelty/helicity
4/6
10.03.2025 19:51 — 👍 2 🔁 0 💬 1 📌 0
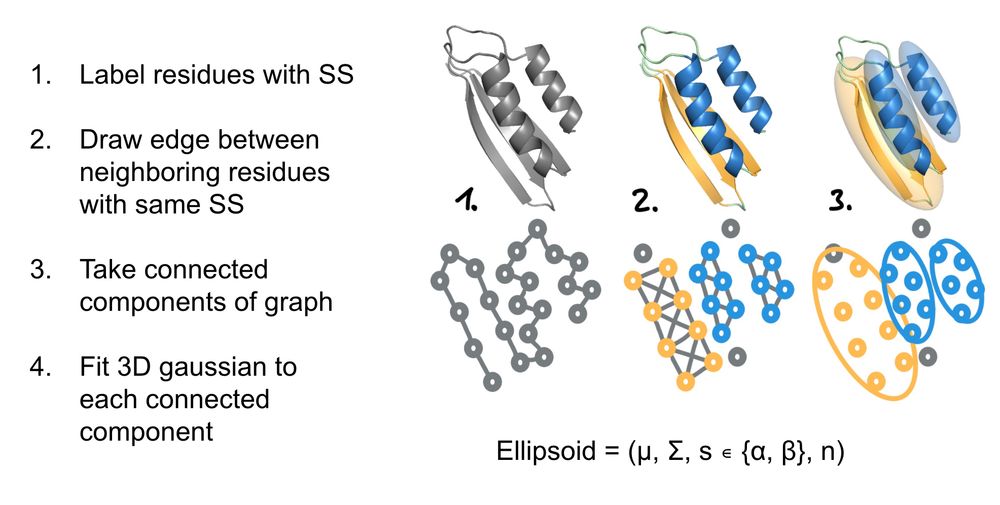
The model just receives the ellipsoids as additional input information.
During training, the ellipsoids are extracted from the training structure (image).
During inference, the ellipsoids can be hand-specified, or come from data, or from our ellipsoid generative model.
3/6
10.03.2025 19:51 — 👍 2 🔁 0 💬 1 📌 0
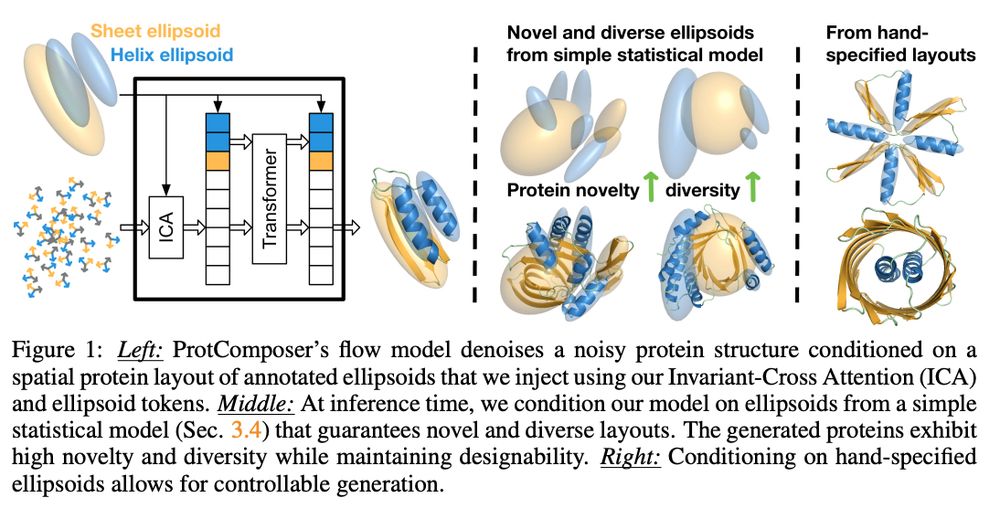
We start with a protein structure generative model (MultiFlow) and finetune it to adhere to 3D ellipsoid layouts. The architecture is modified to process additional ellipsoid tokens and with an attention mechanism for 3D covariance matrices (that is how we represent the ellipsoids).
2/6
10.03.2025 19:51 — 👍 2 🔁 0 💬 1 📌 0
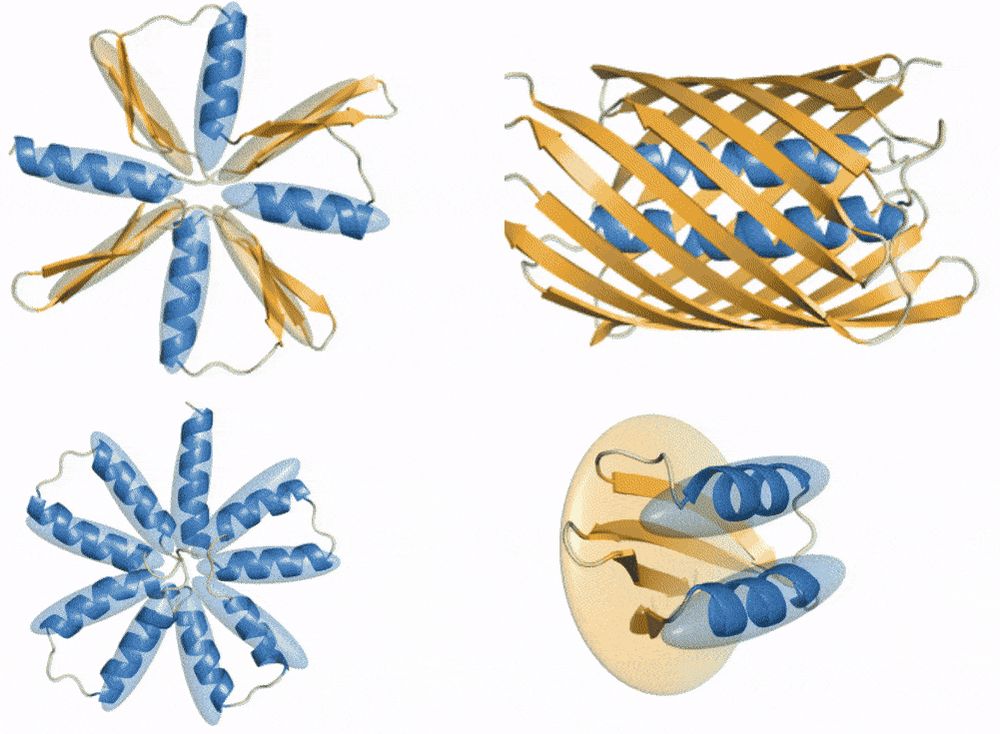
New paper (and #ICLR2025 Oral :)):
ProtComposer: Compositional Protein Structure Generation with 3D Ellipsoids arxiv.org/abs/2503.05025
Condition on your 3D layout (of ellipsoids) to generate proteins like this or to get better designability/diversity/novelty tradeoffs.
1/6
10.03.2025 19:51 — 👍 18 🔁 5 💬 1 📌 1
@garykbrixi.bsky.social @brianhie.bsky.social @patrickhsu.bsky.social @genophoria.bsky.social @anthonybcosta.bsky.social @amyxlu.bsky.social
09.03.2025 17:14 — 👍 0 🔁 0 💬 0 📌 0
personalized cancer immunotherapy = genomics + immunology + machine learning + oncology
(pirl.unc.edu)
Principal Research Scientist at NVIDIA | Former Physicist | Deep Generative Learning | https://karstenkreis.github.io/
Opinions are my own.
Assistant Professor at Stanford Statistics and Stanford Data Science | Previously postdoc at UW Institute for Protein Design and Columbia. PhD from MIT.
Dpt. for #MolecularSociology @mpibp.bsky.social
https://www.biophys.mpg.de/molecular-sociology
Computational chemist/structural bioinformatician working on improving molecular simulation at MRC Laboratory of Molecular Biology. jgreener64.github.io
Chief Science and Strategy Officer, openRxiv. Co-Founder, bioRxiv and medRxiv.
Professor at the NYU School of Medicine (https://yanailab.org/). Co-founder and Director of the Night Science Institute (https://night-science.org/). Co-host of the 'Night Science Podcast' https://podcasts.apple.com/us/podcast/night-science/id1563415749
Head of Sci/cofounder at futurehouse.org. Prof of chem eng at UofR (on sabbatical). Automating science with AI and robots in biology. Corvid enthusiast
machine learning drug discovery scientist @ iambic tx | neuralplexer developer
computational biophysicist, glycoscientist, feminist, jewish, lover of sufjan stevens. she/they 🏳️🌈 ✡︎
📍 sf/nyc
Generative models for protein design.
Group leader at CRG
PhD Candidate in Computational Biology @ University of Pittsburgh. Working on deep generative models for molecular structure. iandunn.io
Promoting fundamental research on advanced computational methods
Genomics, Machine Learning, Statistics, Big Data and Football (Soccer, GGMU)
Co-founder & CSO @Entalpic | AI for materials discovery | PhD in Graph ML, ex-Mila, ex-Amazon | #AIforClimateChange, #AI4Science
🏳️🌈 NVIDIA & Duke. Was Allianz, VantAI, TUM. BioCS+ML dude.
Lab page: https://machine.learning.bio
GScholar: https://scholar.google.com/citations?user=4q0fNGAAAAAJ
MLing biomolecules en route to structural systems biology. Asst Prof of Systems Biology @Columbia. Prev. @Harvard SysBio; @Stanford Genetics, Stats.
Scientist. Christ-follower. Prof @UPenn. Director of Folding@home. Legaly blind. Father.
Director, Max Planck Institute for Intelligent Systems; Chief Scientist Meshcapade; Speaker, Cyber Valley.
Building 3D humans.
https://ps.is.mpg.de/person/black
https://meshcapade.com/
https://scholar.google.com/citations?user=6NjbexEAAAAJ&hl=en&oi=ao
Working towards the safe development of AI for the benefit of all at Université de Montréal, LawZero and Mila.
A.M. Turing Award Recipient and most-cited AI researcher.
https://lawzero.org/en
https://yoshuabengio.org/profile/




























