
Structural architecture of the human long non-coding RNA-PAAN, as a potential target for anti-influenza drug development
www.sciencedirect.com/science/arti...
Really liked the paper but I found it interesting that the word "alignment" only appeared once, in references 🤔
27.09.2025 10:04 — 👍 2 🔁 0 💬 0 📌 0

High-throughput competitive binding assay for targeting RNA tertiary structures with small molecules: application to pseudoknots and G-quadruplexes academic.oup.com/nar/article/...
12.09.2025 18:29 — 👍 3 🔁 1 💬 1 📌 0
Bad RNA 2D → bad 3D — unless the software knows when to say “no thanks.” Can’t wait to read more! 👀
02.07.2025 08:42 — 👍 0 🔁 0 💬 0 📌 0

RNA Resources Project Leader
About the team/job We are recruiting a Project Leader to spearhead the development of the RNAcentral and Rfam databases. Rfam and RNAcentral are key resources for RNA Biology that serve tens of thousa...
I used to lead this team and genuinely loved it: @rnacentral.bsky.social & @rfamdb.bsky.social are hiring a new Project Leader
If you care about RNA, open data, and the future of RNA therapeutics + AI, this is a rare and impactful role in bioinformatics
embl.wd103.myworkdayjobs.com/en-US/EMBL/j...
06.06.2025 10:01 — 👍 1 🔁 0 💬 0 📌 0
NIH funding supporting the HMMER and Infernal software projects has been terminated. NIH states that our work, as well as all other federally funded research at Harvard, is of no benefit to the US.
22.05.2025 12:42 — 👍 287 🔁 232 💬 37 📌 46
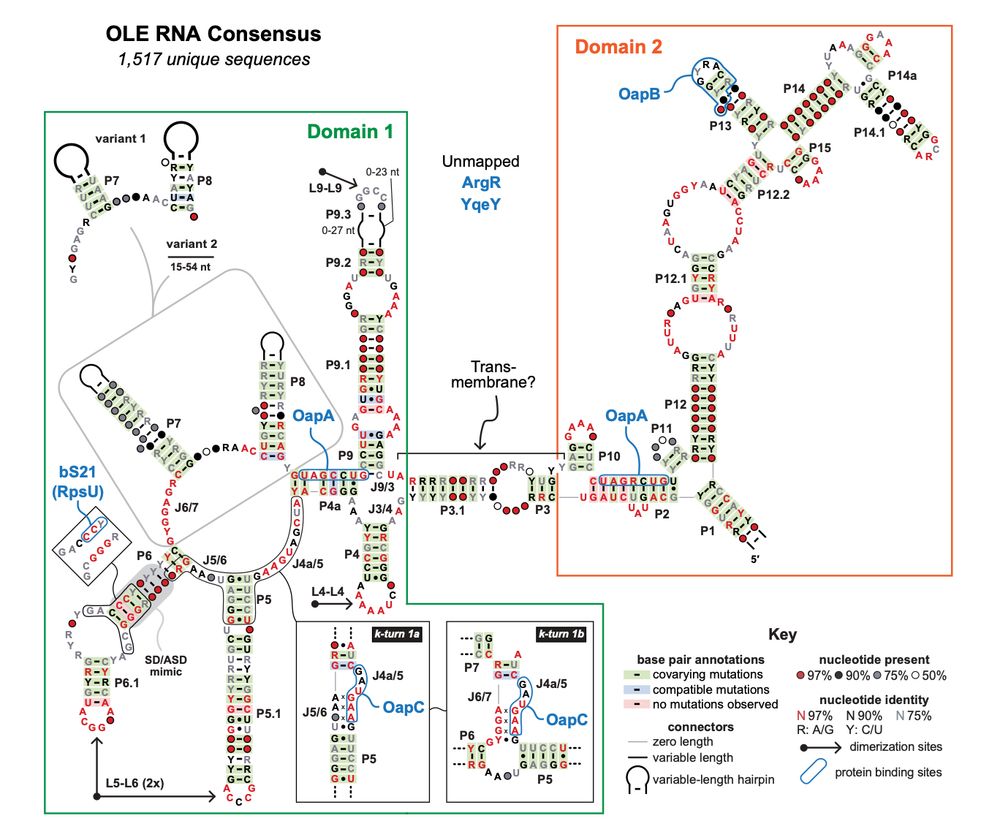
🤯 Implications for OLE RNA as a natural integral membrane RNA from Ron Breaker's lab rnajournal.cshlp.org/content/earl...
23.05.2025 20:21 — 👍 34 🔁 12 💬 1 📌 3
Ensembl release 114 is now out! We have now updated human data to MANE v1.4, new and exciting rodents, livestock, plants and insect genome 🐀 🐈⬛ 🪲, lots of species with variation data (@evarchive) and much more!
More details on our blog (tinyurl.com/3v7cz2sm)%22
#Genomes #bioinformatics
07.05.2025 14:58 — 👍 18 🔁 6 💬 1 📌 0

Quantum algorithm for identifying RNA 3D motifs by processing RNA secondary structure graphs

Figure 1

Figure 2

Figure 3
Quantum algorithm for identifying RNA 3D motifs by processing RNA secondary structure graphs [new]
RNA struct. analysis via QC; encodes base interact. for motifs & drug targets.
29.04.2025 14:29 — 👍 1 🔁 1 💬 0 📌 0

MAFin: Motif Detection in Multiple Alignment Files
AbstractMotivation. Whole Genome and Proteome Alignments, represented by the Multiple Alignment File (MAF) format, have become a standard approach in compa
MAFin: Motif Detection in Multiple Alignment Files | Bioinformatics | Oxford Academic https://academic.oup.com/bioinformatics/advance-article/doi/10.1093/bioinformatics/btaf125/8086993?rss=1&login=false
20.03.2025 02:19 — 👍 1 🔁 1 💬 0 📌 0
It's difficult to counter the basic challenges of RNA 3D -- more torsion angles, magnitudes fewer (unique) PDB structures & I'd add, it's more difficult to build good alignments!
06.05.2025 05:24 — 👍 12 🔁 6 💬 1 📌 0

The crux of rigorously characterizing #IRES:
Kathrin Leppek @katleppek.bsky.social, Maria Barna and colleagues compile a toolbox of approaches to study internal #ribosome entry sites in mammalian cells and embryonic tissues
www.embopress.org/doi/full/10....
13.03.2025 14:03 — 👍 15 🔁 7 💬 0 📌 1
Cool stuff: DMS-FIRST-seq generates high-resolution, accurate RNA structures by coupling chemical probing with nanopore direct RNA sequencing
28.04.2025 12:47 — 👍 5 🔁 2 💬 1 📌 0

rnahub.og
A webserver to generate RNA alignments with secondary structure assessment
www.biorxiv.org/content/10.1...
16.03.2025 12:24 — 👍 30 🔁 11 💬 3 📌 0

openRxiv has arrived!
We’re thrilled to announce the launch of openRxiv as an independent, researcher-led nonprofit to oversee bioRxiv and medRxiv, the world’s leading preprint servers for life and health sciences.
openrxiv.org/introducing-...
#openRxiv #OpenScience #Preprints #bioRxiv #medRxiv
11.03.2025 13:18 — 👍 407 🔁 232 💬 3 📌 19

Lab-in-the-loop therapeutic antibody design with deep learning

Figure 1

Figure 2

Figure 3
Lab-in-the-loop therapeutic antibody design with deep learning [new]
Automated Ab design using iterative ML, property prediction, & lab experiments, yielding improved binders against key targets.
25.02.2025 01:27 — 👍 2 🔁 1 💬 0 📌 0

Micromix: web infrastructure for visualizing and remixing microbial ‘omics data
Abstract. Micromix is a flexible web platform for sharing and integrating microbial omics data, including RNA sequencing and transposon-insertion sequencin
Presenting a new flexible web platform for sharing and integrating microbial omics data, including RNA-seq and transposon-insertion sequencing.
Micromix: web infrastructure for visualizing and remixing microbial ‘omics data doi.org/10.1093/giga...
19.02.2025 09:36 — 👍 8 🔁 3 💬 0 📌 0
#RNA rules
26.02.2025 18:54 — 👍 15 🔁 3 💬 0 📌 1
Many @ebi.embl.org resources rely on R2DT for RNA visualisation - huge thanks to everyone for their amazing work! 🙌 🧬
10.02.2025 14:43 — 👍 2 🔁 0 💬 0 📌 0

Last but not least, a huge thanks to the amazing R2DT team! Shoutout to teams at @ebi.embl.org, @pdbeurope.bsky.social, @rnacentral.bsky.social, @rfamdb.bsky.social and many more for their great contributions! 🌟
10.02.2025 14:38 — 👍 5 🔁 0 💬 0 📌 0

R2DT · Visualise RNA secondary structure
R2DT is a tool for visualising RNA secondary structure.
R2DT can be used on the web, through an API, in Jupyter notebooks, or locally with #Docker 🚀
Get started at r2dt.bio and reach out with any questions!
#RNA #Bioinformatics #dataviz
10.02.2025 14:38 — 👍 3 🔁 0 💬 1 📌 0
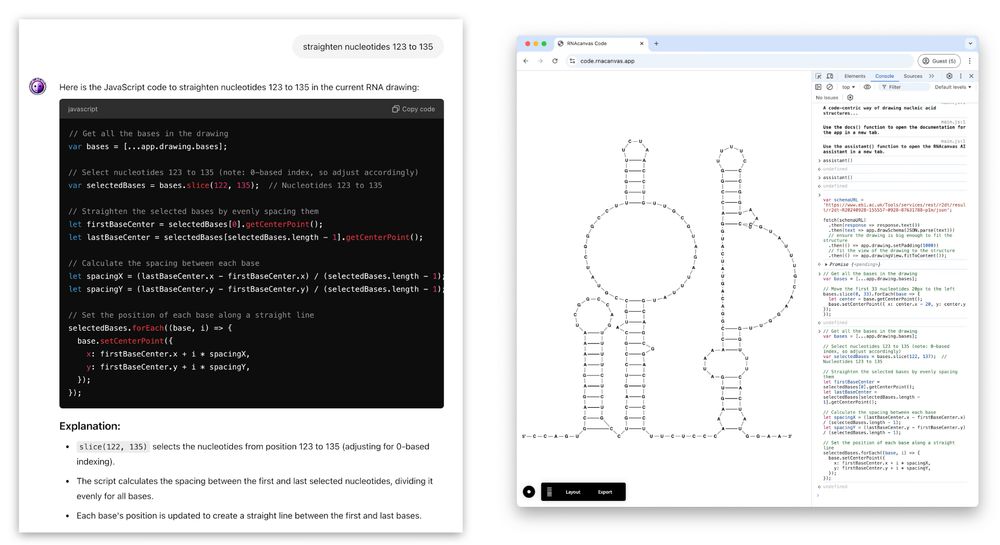
🤖 Simply type, ‘Straighten nucleotides 123 to 135’ – RNAcanvas Code converts your commands to instructions using a custom ChatGPT ✨
10.02.2025 14:38 — 👍 2 🔁 0 💬 1 📌 0
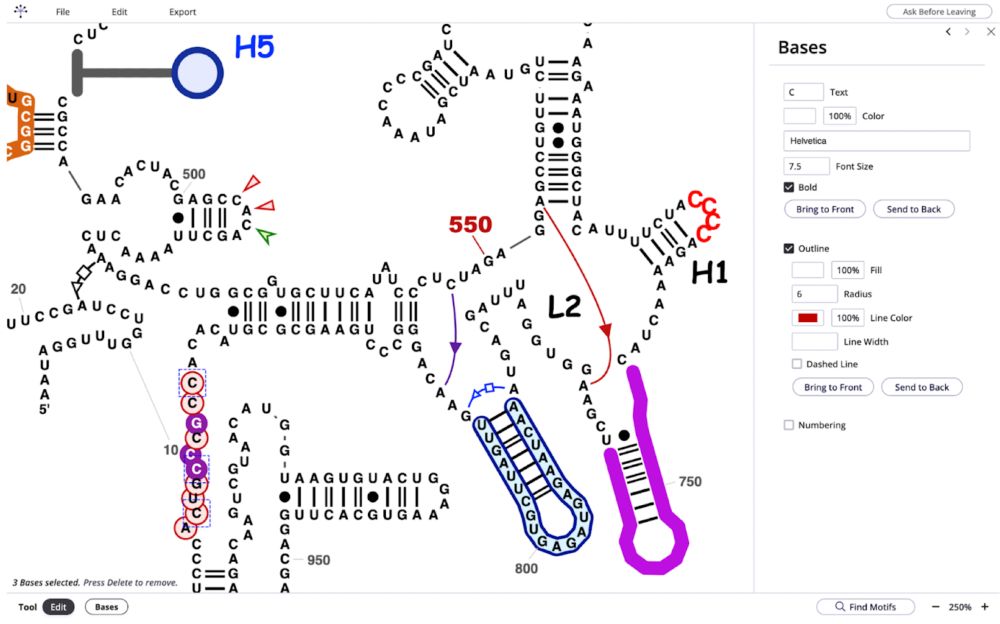
The R2DT diagrams can be edited online using 𝐑𝐍𝐀-𝐬𝐩𝐞𝐜𝐢𝐟𝐢𝐜 editors and the tweaked layouts can be imported back into R2DT 🎨🔄
10.02.2025 14:38 — 👍 2 🔁 0 💬 1 📌 0
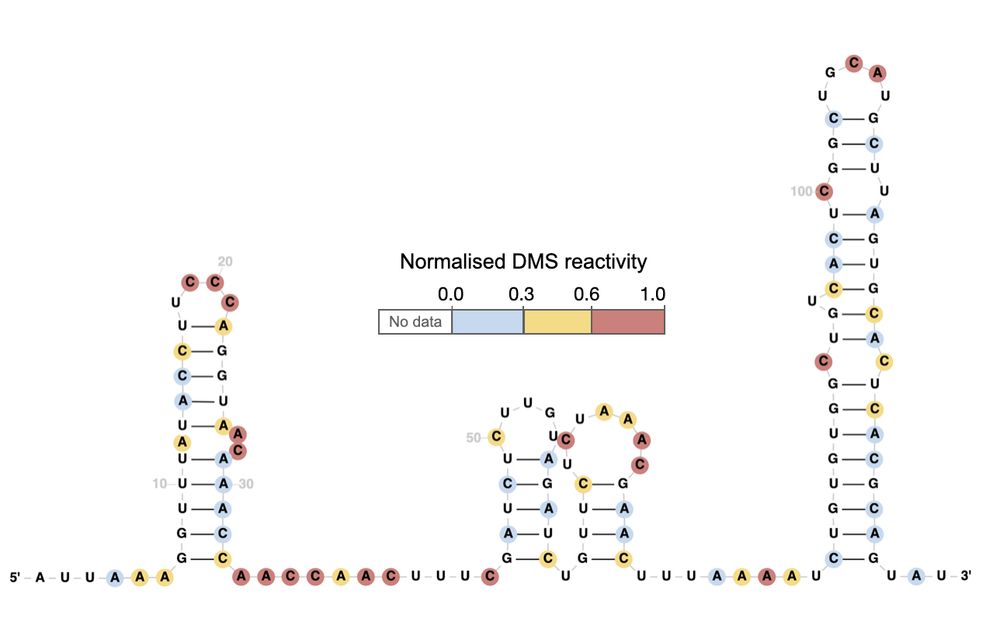
R2DT can also visualise 𝐚𝐧𝐲 𝐝𝐚𝐭𝐚 overlaid on top of RNA structures. Think reactivities, conservation, SNPs, 3D distances, or anything else! 🧬✨
10.02.2025 14:38 — 👍 1 🔁 0 💬 1 📌 0
RNA Resources Project Leader at @ebi.embl.org RNAcentral, Rfam
Community initiative for biomedical MCP servers 🤖🧬
Automatically shares new MCP servers from the Registry
Find out more at: https://biocontext.ai
Post-Doctoral Fellow at University of Bern @unibe.ch working on post-transcriptional gene regulation
#Bioinformatician
#RNAstructure
#RNAbiology
The Earth BioGenome Project (EBP), a moonshot for biology, aims to sequence, catalog, and characterize the genomes of all of Earth's eukaryotic biodiversity over a period of ten years.
🌲Keep up with all EBP updates: https://linktr.ee/earthbiogenomeproject
Project Lead at @ebi.embl.org working on Bioinformatics Tools and APIs.
Eterna empowers citizen scientists to help advance medicine through molecular design. Solve puzzles using RNAs, the tiny molecules at the heart of every cell.
Website: https://eternagame.org/
RNA, viruses, and everything in between | Postdoc in Doudna and Cate Labs @UCBerkeley
Postdoctoral fellow at @miskalab.bsky.social @cambridgeuni.bsky.social.
Nanomedicine•Oligonucleotide therapy•ncRNA•cancer
🧬🔬😊
The Alliance of Genome Resources supports biological science via data integration, standardization of data models and interfaces, and unified outreach.
Find us at https://www.alliancegenome.org.
Curating and cataloging the world's data for the fruit fly Drosophila melanogaster into our database at http://flybase.org. Aiding discovery since 1992.
Curator, nomenclature advisor and group coordinator at FlyBase (Cambridge UK). Special interest in Drosophila metabolic pathways, enzymes, ncRNAs and gene groups.
Professor in bioinformatics, Stockholm University. Protein structure lover ( interactions predictions, evolution ..). Using machine learning as a part of AI for Sciences for halv my life. In addition to succén, I sometimes rants about sailing or skiing.
Our team studies #Rloop & #RNA 🧬 with #biochemistry #cryoEM 🔬❄️ #crystallography💎 to understand how they control life! Views from Charles
https://biochem.emory.edu/bou-nader-lab/
Associate Professor @ Big Data Institute, University of Oxford
2024 Lister Institute Fellow
genomics | rare disease | gene regulation | genetic therapies
https://rarediseasegenomics.org/
(field) hockey player | cyclist | hiker
The European Molecular Biology Laboratory drives visionary basic research and technology development in the life sciences. www.embl.org
InterPro provides functional analysis of proteins by classifying them into families and predicting domains and important sites.
ebi.ac.uk/interpro/
We develop (non-coding) RNA therapeutics for incurable diseases using genomics and bioinformatics. PI: Rory Johnson





























