Amen 🙏🏻 🤲🏻
A wise person once told me "all antibodies are non-specific until proven otherwise" I was just telling this to my students today 🤯
Amen 🙏🏻 🤲🏻
A wise person once told me "all antibodies are non-specific until proven otherwise" I was just telling this to my students today 🤯
Excited to share our preprint exploring the structural, biochemical and in vivo properties of DRT7/UG10 reverse transcriptases which produce dsDNA using their primase and polymerase activities and provide broad-spectrum antiphage defense! www.biorxiv.org/content/10.6...
25.02.2026 11:33 — 👍 26 🔁 10 💬 2 📌 0very unusual - a tRNA regulated anion channel
18.12.2025 17:26 — 👍 57 🔁 23 💬 2 📌 2
Last X-Mas, the ribosome gave you methionine,
but the very next day, MetAP took it away.
This year, to save histones from tears,
NatD gives you an acetyl group. ⭐️
Explore our latest paper with the Deuerling lab @uni-konstanz.de and Shu-ou Chan lab @caltech.edu!
www.science.org/doi/10.1126/...

@science.org Chromatin buffers torsional stress during transcription | Science www.science.org/doi/10.1126/...
19.12.2025 22:31 — 👍 32 🔁 11 💬 0 📌 0It’s out! Using cryo-ET in Dicty cells, we take a fresh in situ look at vaults. Surprisingly, we uncover vaults associated with ER and NE membranes, and find that many vaults enclose ribosomes in defined orientations, opening new avenues to their cellular function! www.biorxiv.org/lookup/conte...
16.12.2025 08:17 — 👍 106 🔁 39 💬 2 📌 5
Our latest paper on the molecular mechanisms of XPF-ERCC1 recruitment to SLX4-dependent DNA repair pathways has been published! Congratulations to first author Junjie Feng, and a heartfelt thank you to all our collaborators for their contributions to this study!
www.nature.com/articles/s41...

The dark proteome: translation from noncanonical open reading frames
1⃣5'-UTR 2⃣3'-UTR small ORF/sORF
3⃣out-of-frame 4⃣In-frame overlapping ORF
Translating micropeptide <100aa, proteins distinct from canonical ORFs, or isoforms
#TrendsCellBio 2022
www.sciencedirect.com/science/arti...
✨New preprint!
🧵1/4 Excited to share our work on AI-guided design of minimal RNA-guided nucleases. Amazing work by @petrskopintsev.bsky.social @isabelesain.bsky.social @evandeturk.bsky.social et al!
Multi-lab collaboration @banfieldlab.bsky.social @jhdcate.bsky.social @jacobsenucla.bsky.social🧬
🔗👇

Colibactin-DNA interstrand crosslinks structure reveals DNA-damaging acitivity of colibactin that is linked to colorectal cancer #MicroSky www.science.org/doi/10.1126/...
04.12.2025 22:07 — 👍 21 🔁 10 💬 1 📌 1
❄️ December RNA Modifications Newsletter is out! ❄️
rnamodifupdates.substack.com/p/rna-modifi...
❄️🔥RNA modifs = winter’s hottest adaptation toolkit. 🦠
#Ribosome #RNAbiology
From @kylecottrell.bsky.social lab
rnajournal.cshlp.org/content/31/1...
So happy to see this work out! Was such a pleasure to co-lead this effort with Erin. Do you like viral immune evasion, and using protein structure to study immune antagonists? Give it a read!
25.11.2025 21:34 — 👍 17 🔁 8 💬 0 📌 0
Congrats to @chgjohnston.bsky.social , Andreani, Radicella et al.!
Elegant PNAS paper on the ComEC/ComFA/ComFC complex in bacterial transformation. AF3 predictions validated by clever mutagenesis. We tried for years to solve this by cryo-EM/crystallography - never succeeded.
Happy to share that my PhD project is finally published!🪱✨
Selfish genes are found across the tree of life. They can disrupt inheritance patterns and at the same time act as units for molecular innovation. Here we tried to answer one big question: how do selfish genes emerge in the first place?

How does messenger RNA (mRNA) get out of the nucleus to become a protein? Eukaryotic mRNA is packaged, exported, and then translated in the cytoplasm. But how do these steps work? And what are open questions? Check out our new review for our take: www.annualreviews.org/content/jour... (1/3)
21.11.2025 17:36 — 👍 122 🔁 52 💬 1 📌 3
New work from the Green and Beckmann labs on how ZAK recognizes collided ribosomes.
www.nature.com/articles/s41...

To probe gene-scale chromatin physics, we built 96-mer (20 kb) arrays with defined histone marks. Combining single-molecule tracking, AFM imaging, and developing in vitro Hi-C, we saw how specific modifications dictate chromatin structure and dynamics. www.science.org/doi/10.1126/...
20.11.2025 07:46 — 👍 60 🔁 21 💬 5 📌 1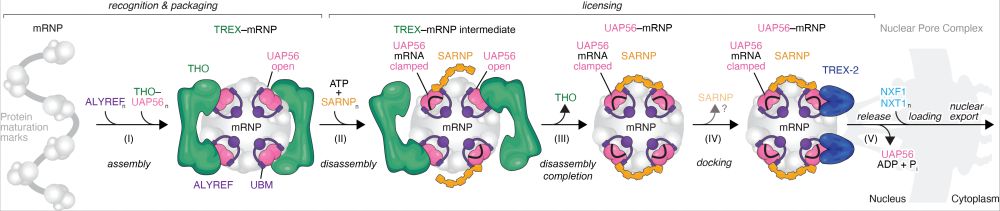
Finally out in @nature.com! We uncovered a mechanistic framework for a general and conserved mRNA nuclear export pathway. www.nature.com/articles/s41.... 1/
19.11.2025 23:21 — 👍 106 🔁 42 💬 3 📌 1
Out today, our take on 6-methyladenine #6mA evolution in Eukaryotes @natgenet.nature.com. We asked a simple question, is really DNA 6mA common across the eukaryotes? The answer is "yes" if you're a unicellular eukaryote 🦠, not so if you're multicellular 🐝🌱🍄. www.nature.com/articles/s41... 1/9
18.11.2025 12:00 — 👍 164 🔁 85 💬 7 📌 6
Structural insights into the chaperone role of uncharged tRNA(Arg/Gln) in viral RNAP assembly #RNASky 🧪 www.nature.com/articles/s41...
15.11.2025 21:44 — 👍 8 🔁 5 💬 0 📌 0
Excited to share our new preprint in collaboration with Ahmet Yildiz's lab. Check out how our team uncovers a novel binding footprint and motor regulation mechanism for MAP9 Congrats to Burak Cetin and @aryantaheri.bsky.social
www.biorxiv.org/content/10.1...

Very happy to share our collaborative project on FAM118 proteins - noncanonical sirtuins that form filaments and process NAD in human and other vertebrate cells.
17.11.2025 11:37 — 👍 75 🔁 32 💬 2 📌 4
Thank you to our sponsors for the GA RNA Salon @rnasociety.bsky.social and Lexogen- we hosted Dr. Francesca Storici for our annual symposium yesterday @dunhamlab.bsky.social @ghaleilab.bsky.social
15.11.2025 14:22 — 👍 19 🔁 5 💬 0 📌 0
I’m very happy to share Zhong Han’s beautiful work on the biochemical function of Senataxin, encoded by a gene that is mutated in rare and early-disabling neurodegenerative diseases. Turns out it rescues backtracked RNA polymerase II during early transcription! www.sciencedirect.com/science/arti...
12.11.2025 17:36 — 👍 19 🔁 8 💬 0 📌 0
Thrilled to share that the final piece of my PhD work is now on bioRxiv! biorxiv.org/content/10.1... With support from @nvidia and the @NSF, we used AlphaFold to screen 1.6M+ protein pairs, revealing thousands of potential novel PPIs. All data can be viewed at predictomes.org/hp
12.11.2025 21:26 — 👍 163 🔁 67 💬 5 📌 4
Regulated decay of microRNAs plays a critical role in controlling body size in mammals! Check out our new paper in @genesdev.bsky.social and see thread previously posted with our pre-print 👇 for more info. Congrats to Collette LaVigne, Jaeil Han, and all authors!
genesdev.cshlp.org/cgi/content/...

Very proud of our recently published work in @narjournal.bsky.social led by senior PhD candidate Julia Warrick. #RNA
academic.oup.com/nar/article/...

Tremendous paper from Narry Kim’s lab identifying a sequence element that recruits cellular factors to exogenous mRNAs to ‘rejuvenate’ their poly(A) tails leading to a huge boost in output. Significant implications for therapeutics. Now, I wonder what the factor X is…
www.nature.com/articles/s41...

New work from Tahirov x Lim collaboration! @qixianghe.bsky.social from my lab contributed the cryo-EM structures for this work. We are excited to help explain how anti-HSV drugs work and to guide their future development.
www.science.org/doi/10.1126/...
@unmc.bsky.social @uwbiochem.bsky.social