


🚀 InterPro 108.0 is live!
✨ Updated to Pfam 38.1 and NCBIFam 18.0
✨ 1,911 InterPro entries
Explore the latest annotations and discover new protein insights.
Explore it now: www.ebi.ac.uk/interpro/
@interprodb.bsky.social
InterPro provides functional analysis of proteins by classifying them into families and predicting domains and important sites. ebi.ac.uk/interpro/



🚀 InterPro 108.0 is live!
✨ Updated to Pfam 38.1 and NCBIFam 18.0
✨ 1,911 InterPro entries
Explore the latest annotations and discover new protein insights.
Explore it now: www.ebi.ac.uk/interpro/



🚀InterPro 107.0 is live!
✨Updated to Pfam 38.0
✨1,068 new InterPro entries
Explore the latest annotations and discover new protein insights.
Explore it now: interpro.ebi.ac.uk

InterPro page with a list of the quinoa sequences in UniProtKB that belong to the HSP70 family.
Quinoa thrives at 4,000m altitude where many crops struggle. Heat Shock Proteins helps protect plant cells from environmental stress. These molecular mechanisms in traditional crops may inform climate-resilient agriculture strategies. 💻🧪
More info: proteinswebteam.github.io/interpro-blo...
Interested in studying Parkinson's disease? Have a look at our blog post below
17.09.2025 10:24 — 👍 2 🔁 1 💬 0 📌 0📢 Happy to announce that Ensembl 115 and Ensembl Genomes 62 are out! This release features ~120000 new protein coding transcripts in human GRCh38, 2 new cattle, 7 new plants and 20 new metazoa! 🧬🐮 🫛🪲
Read more here ➡️ zurl.co/poyXM
Try it now on your favorite InterPro entry! www.ebi.ac.uk/interpro/ent...
27.08.2025 15:55 — 👍 0 🔁 0 💬 0 📌 0Click any segment to:
✨List matching proteins
✨Download FASTA files
✨Jump to the taxonomy entry for deeper insights
Control the depth and display mode—view by number of sequences or number of species.
27.08.2025 15:55 — 👍 0 🔁 0 💬 1 📌 0It maps protein occurrences from broad kingdoms down to specific species. See how widely a domain or site appears across life.
27.08.2025 15:55 — 👍 0 🔁 0 💬 1 📌 0Feature Spotlight: Taxonomy Sunburst
Discover the taxonomic breadth of your InterPro entry at a glance with our dynamic sunburst chart! 🕸️ 💻🧪🧬

InterProScan 6 (beta) Nextflow - Containerised - Reproducible outputs Start with FASTA file InterProScan workflow: Annotate proteins (and nucleotides) with families, domains & sites Select analyses (e.g. Pfam, PANTHER, CDD, NCBIFAM...) Optional: GO and pathway annotations Output formats: JSON, TSV, XML, GFF3
InterProScan 6 (beta) is here 🎉
A Nextflow-powered, containerised overhaul for scalable, reproducible protein annotation—decoupled software/data, automated data handling, and smoother runs across HPC and cloud.
Feedback welcome!
github.com/ebi-pf-team/...
💻🧪🧬

Poster A-098: Pfam database in 2025 – towards expanding protein families coverage Nicole Morveli Flores Session A: 21/07/2025 10:00 – 11:20 and 16:00 – 16:40 BST Image credit: BioImage Archive accession S-BSST584. Design credit: Karen Arnott/EMBL-EBI
InterProScan 6: a modern large-scale protein function annotation pipeline Matthias Blum 21/07/2025 14:20 Room 03A Image credit: BioImage Archive accession S-BSST584. Design credit: Karen Arnott/EMBL-EBI

Poster A-205: Carbohydrate active enzymes in Pectobacteriaceae: coevolving enzyme sets and host adaptation Emma Hobbs Session A: 21/07/2025 10:00 – 11:20 and 16:00 – 16:40 BST Image credit: BioImage Archive accession S-BSST584. Design credit: Karen Arnott/EMBL-EBI

Accelerating protein family classification in InterPro with AI innovations Matthias Blum 22/07/2025 16:40 Room 12 Image credit: BioImage Archive accession S-BSST584. Design credit: Karen Arnott/EMBL-EBI
🧬 The InterPro/Pfam team will be presenting at #ISMB2025 next week in Liverpool!
🎤 Multiple talks & 📊 posters on protein analysis & classification
💻🧪

The InterPro/Pfam team members attending ISMB/ECCB 2025: Alex Bateman, Emma Hobbs, Matthias Blum, Nicole Morvelli, Antonina Andreeva Image credit: BioImage Archive accession S-BSST584. Design credit: Karen Arnott/EMBL-EBI
🧬 The InterPro/Pfam team will be presenting at #ISMB2025 next week in Liverpool!
Join us July 20-24 at the world's largest #bioinformatics conference!
#ISMBECCB2025 #ProteinAnalysis #ComputationalBiology
💻🧪

JOBIM 2025 logo 8 to 11th July 2025 Bordeaux
🧬✨ Excited to present at @jobim2025.bsky.social this week in Bordeaux!
🎤 Talk 08/07 16:50, amphi E: Pfam: embracing AI/ML by @typhainepl.bsky.social
📊 Poster #97: InterPro: Accelerating Protein Annotation with AI by @matthiasblum.bsky.social
💻🧪
Level up your protein prediction skills with the updated EMBL-EBI Training AlphaFold practical course.
Now featuring #AlphaFold Server and AlphaFold 3. Start your training today: www.ebi.ac.uk/training/onl...
AlphaFold, developed by @ebi.embl.org and Google DeepMind.
![Schematic diagram showing taste signal transmission between the tongue (with taste buds that contain taste cells harbouring GPCRs) and brain. Taste GPCRs are activated by specific food compounds, recruit specific G proteins that further induce intracellular calcium release. Adapted from [https://bio.libretexts.org/Bookshelves/Biochemistry/Fundamentals_of_Biochemistry_(Jakubowski_and_Flatt)/Unit_IV_-_Special_Topics/28:_Biosignaling_-_Capstone_Volume_I/28.18:_Signal_Transduction_-_Taste_(Gustation)].](https://cdn.bsky.app/img/feed_thumbnail/plain/did:plc:2b2g3csqntyhbjba6zutidvo/bafkreiarei32po5buevs63s7abazj42rdvx5w7pnszlg2zduhyqlpfqxdu@jpeg)
Schematic diagram showing taste signal transmission between the tongue (with taste buds that contain taste cells harbouring GPCRs) and brain. Taste GPCRs are activated by specific food compounds, recruit specific G proteins that further induce intracellular calcium release. Adapted from [https://bio.libretexts.org/Bookshelves/Biochemistry/Fundamentals_of_Biochemistry_(Jakubowski_and_Flatt)/Unit_IV_-_Special_Topics/28:_Biosignaling_-_Capstone_Volume_I/28.18:_Signal_Transduction_-_Taste_(Gustation)].
Does taste depend on our genes? Discover how variations in taste receptors—from TAS2Rs that gate bitterness to olfactory genes that make cilantro taste soapy—and even pain receptors for spice shape what we love (and hate) on our plates. Read more: proteinswebteam.github.io/interpro-blo... 💻🧪👅🧬
26.06.2025 09:44 — 👍 1 🔁 0 💬 0 📌 0
InterPro 106.0 19th June 2025
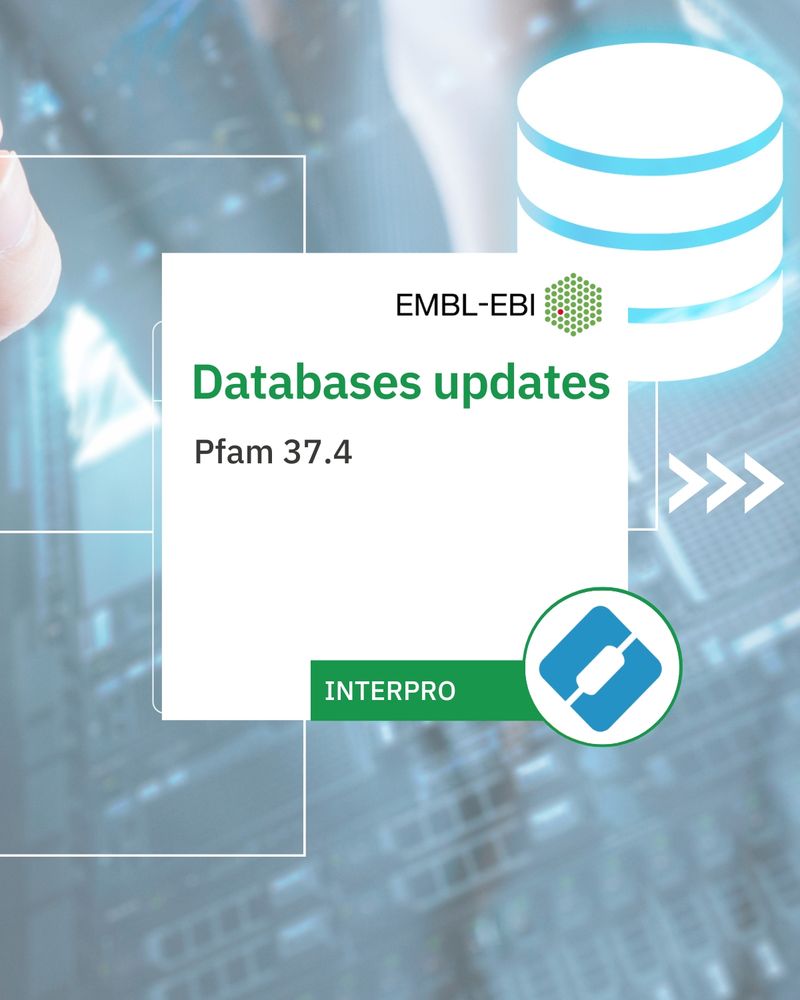
Databases updates: Pfam 37.4

New entries: 683 new entries with: 613 Pfam 78 NCBIfam 30 CDD 4 PANTHER ...
🚀 InterPro 106.0 is now live!
✨ Updated to Pfam 37.4
✨ 683 new entries added
✨ Enhanced protein classification coverage
Explore the latest annotations and discover new protein insights.
Explore it now: interpro.ebi.ac.uk 💻🧪🧬
#Bioinformatics #ProteinScience #Pfam #EMBLEBI
What’s new in the redesigned PDBe entry pages?
🧪 Cleaner layout
🧪 Interactive 3D viewers
🧪 Contextual annotations
🧪 Easier navigation
See for yourself:
wwwdev.ebi.ac.uk/pdbe/entry/p...
#PDBeBeta #Bioinformatics #Biochemistry #Science
InterPro just got one of its biggest updates ever.
Now with 1.8 billion AI-driven protein annotations, AlphaFold-based TED domains, and viral proteins from the Big Fantastic Virus Database (BFVD).
See it all at www.ebi.ac.uk/interpro
🖥️🧬
@interprodb.bsky.social
@martinsteinegger.bsky.social
03.06.2025 12:55 — 👍 0 🔁 0 💬 0 📌 0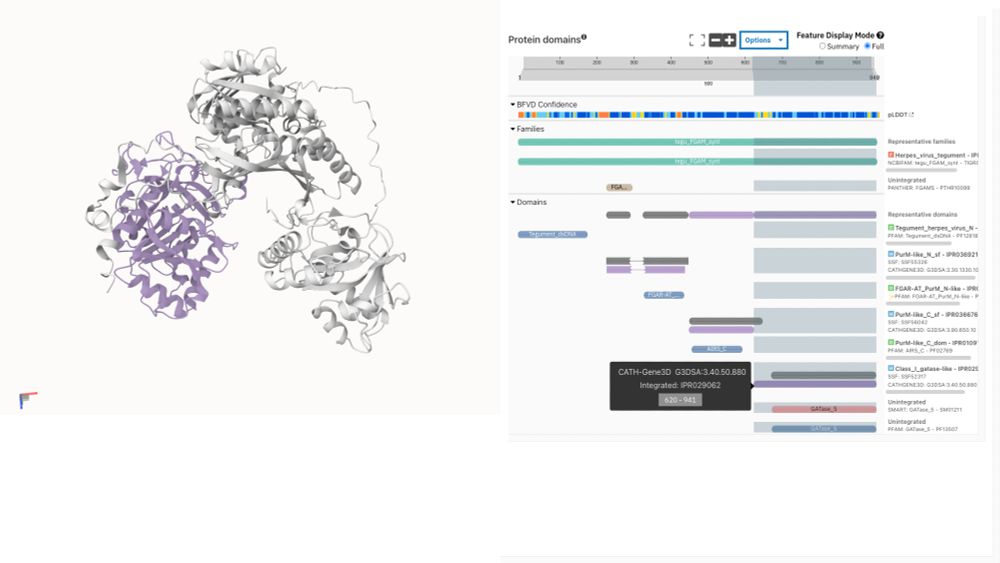
CATH-Gene3D annotation visualised on BFVD structure prediction of UniProt A0A075FDU7
🦠🧬Exciting news! BFVD predicted viral protein structures are now available in @InterProDB!
For the first time, explore InterPro functional annotations directly on 3D structures of viral proteins not covered by AlphaFold DB. This bridges a critical gap in structural virology research.
We are saddened to hear of the passing of Daniel Kahn, the creator of the ProDom database. His pioneering work in protein domain classification laid important groundwork for InterPro and the wider bioinformatics community. We honour his lasting contributions. 🧬🕊️
20.05.2025 08:29 — 👍 2 🔁 0 💬 0 📌 0🧬 MAJOR UPDATE: InterPro-N matches now available in InterPro!
1.8 billion AI-driven protein annotations developed with Google DeepMind are now live.
✨ Look for the sparkles icon to identify them
📊 Options menu lets you view: InterPro only | InterPro-N-only | Stacked
💻🧪
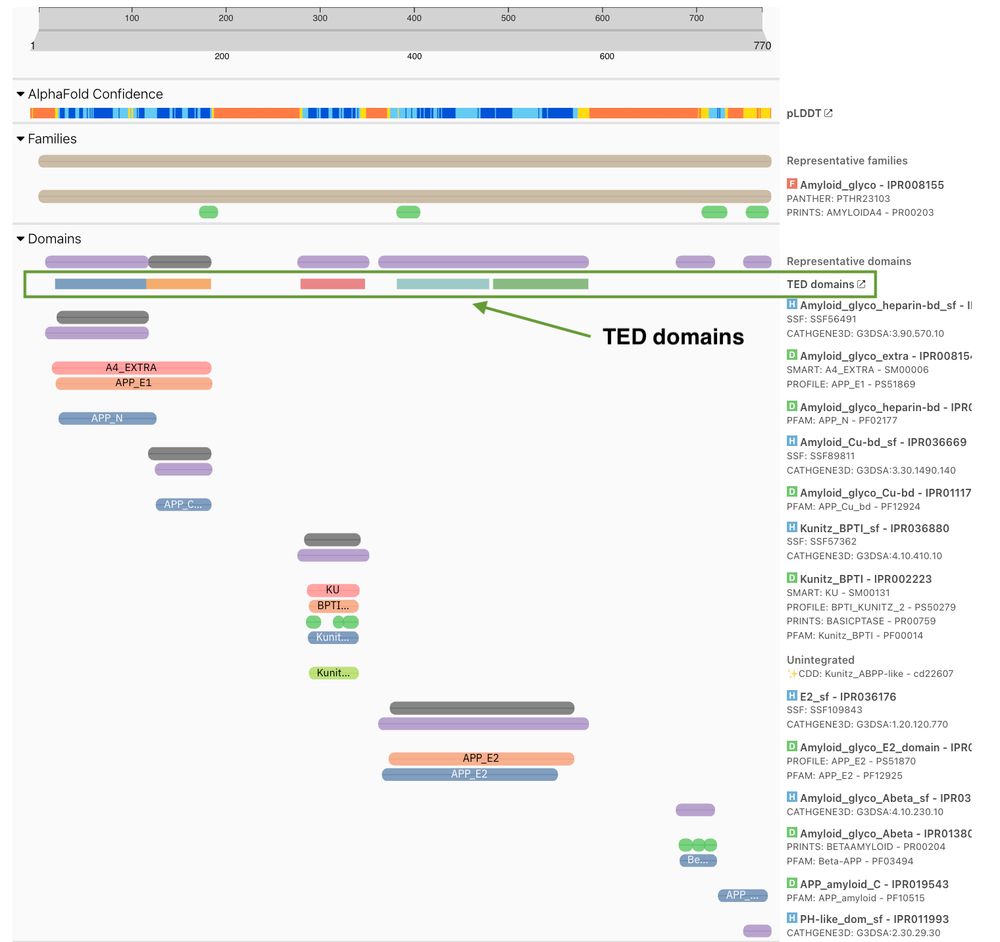
TED domains alongside InterPro annotations for UniProt P05067
🔍 NEW: TED domains in InterPro 105.0!
For the first time, see AlphaFold-derived structural domains directly alongside InterPro annotations in one powerful visualisation. Bridging the sequence-structure divide in protein analysis. 💻🧪
Explore now: www.ebi.ac.uk/interpro/pro...
InterPro 105.0: AI for protein classification InterPro 105.0 is now live. This AI-driven update makes it easier than ever to explore the protein universe
📝 Check out our new blog post highlighting the groundbreaking features in InterPro 105.0!
Learn how InterPro-N, TED domains and viral protein structures are transforming protein classification: ebi.ac.uk/about/news/u...

InterPro 105.0 24th April 2025

Databases updates Pfam 37.3 PROSITE patterns 2025_01 PROSITE profiles 2025_01 HAMAP 2025_01

New entries 342 new entries with: 318 Pfam 40 NCBIfam 12 Prosite profiles 6 PANTHER 5 CDD

New features InterPro-N annotations BFVD structure predictions Simplified structure pages
📢InterPro release 105.0 is live!
✨Update to Pfam 37.3, PROSITE patterns 2025_01
PROSITE profiles 2025_01 and HAMAP 2025_01
✨342 new entries
✨Addition of InterPro-N annotations
✨Addition of BFVD structure predictions from @martinsteinegger.bsky.social's lab
Have a look: ebi.ac.uk/interpro/htt...
💻🧪🆕

LLM generated summaries for protein classification at InterPro
Missed our latest webinar about LLM generated summaries for protein classification at InterPro delivered by
@matthiasblum.bsky.social?
The recording is now available: lnkd.in/ei4fKGXd
Want to use the InterPro API but not sure how to start? Join this upcoming workshop, where @typhainepl.bsky.social will guide you through the fundamentals and practical applications. Discover how to integrate this powerful tool into your workflow! Register now to secure your spot. 💻🧪
02.04.2025 09:36 — 👍 4 🔁 2 💬 0 📌 0
Are you going to ISMB/ECCB 2025 @iscb.bsky.social in Liverpool 🇬🇧? Register for our tutorial on 'Genomic Variant Interpretation & prioritisation for clinical research'. More info: tinyurl.com/ahav9djn
#genomics #bioinformatics

InterProScan Version: 5.73-104.0 6th February 2025
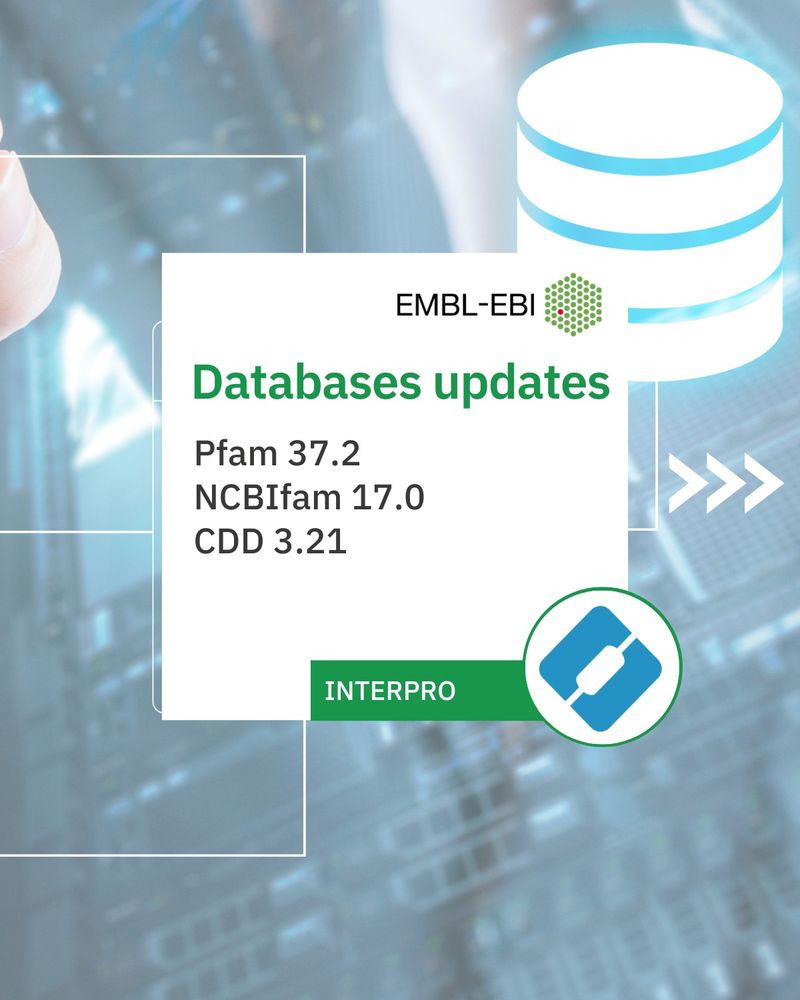
Databases updates: Pfam 37.2 NCBIfam 17.0 CDD 3.21

Ways to access: 1) InterPro website: sequence search 2) Local installation Software download Docker container
🆕 New version of InterProScan: 5.73-104.0
3 ways to use it:
1️⃣ Sequence search on the InterPro website: ebi.ac.uk/interpro/sea...
2️⃣ Local installation: interproscan-docs.readthedocs.io/en/v5/Instal...
3️⃣ Local installation using a container: interproscan-docs.readthedocs.io/en/v5/HowToU...
💻🧪