Honored to be named the Laura and Isaac Perlmutter Endowed Chair in Biochemistry at NYU Grossman School of Medicine. Grateful to my mentors, lab members, and the NYU community—and most of all, to my amazing family for their constant support!
17.10.2025 16:03 — 👍 41 🔁 3 💬 14 📌 0
Thanks Aidan! Very on-point comment, I appreciate it - and you are right. Often, one tries to simplify the system by limiting the number of variables for a single study - but in reality it seems more like an elaborate albeit stochastic ballet dance :)
01.06.2025 13:20 — 👍 1 🔁 0 💬 0 📌 0
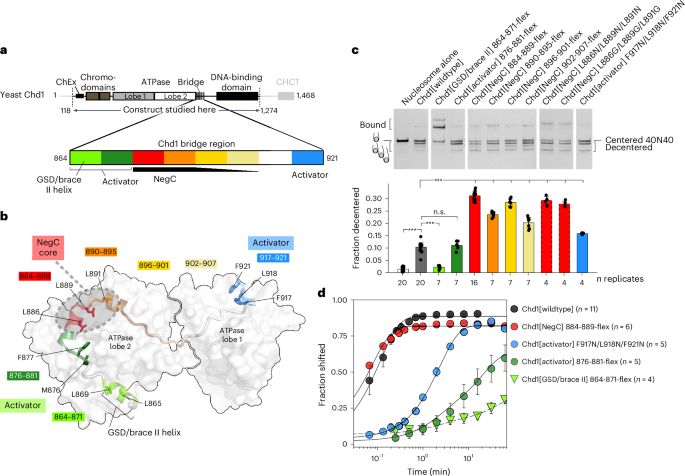
We coupled in depth mutagenesis and biochemistry with high-resolution cryoEM, obtaining maps ranging from 2.37 to 2.9 Å.
This work was done by Ilana Nodelman (Bowman lab) and Heather Folkwein (Armache lab)
Thank you to everyone involved. It was a fun joint-venture!
30.05.2025 15:12 — 👍 6 🔁 1 💬 1 📌 0
Thank you🙏
30.05.2025 15:04 — 👍 0 🔁 0 💬 0 📌 0
This was a highly collaborative work between multiple labs, performed by a talented grad student in the Armache and Murakami labs, Natalie Smith. Thank you to all the authors for their incredible work!
05.02.2025 01:03 — 👍 4 🔁 0 💬 0 📌 0
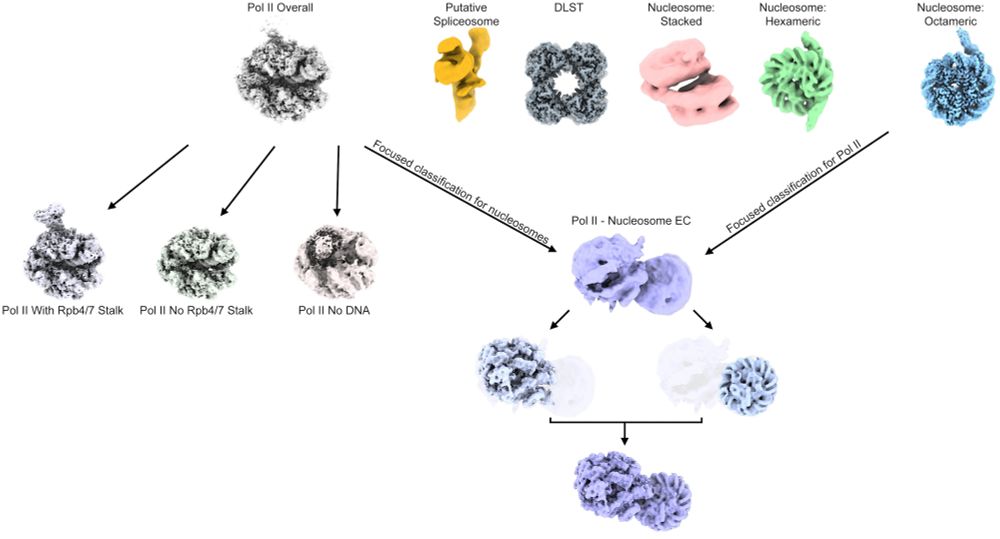
We did a thing... We purified native transcriptional complexes from Drosophila embryos, obtaining +/- stalk Pol II elongation complexes, native nucleosomes, as well as a nucleosome elongation complex. For more details please check out: doi.org/10.1101/2025...
05.02.2025 01:02 — 👍 56 🔁 20 💬 1 📌 1
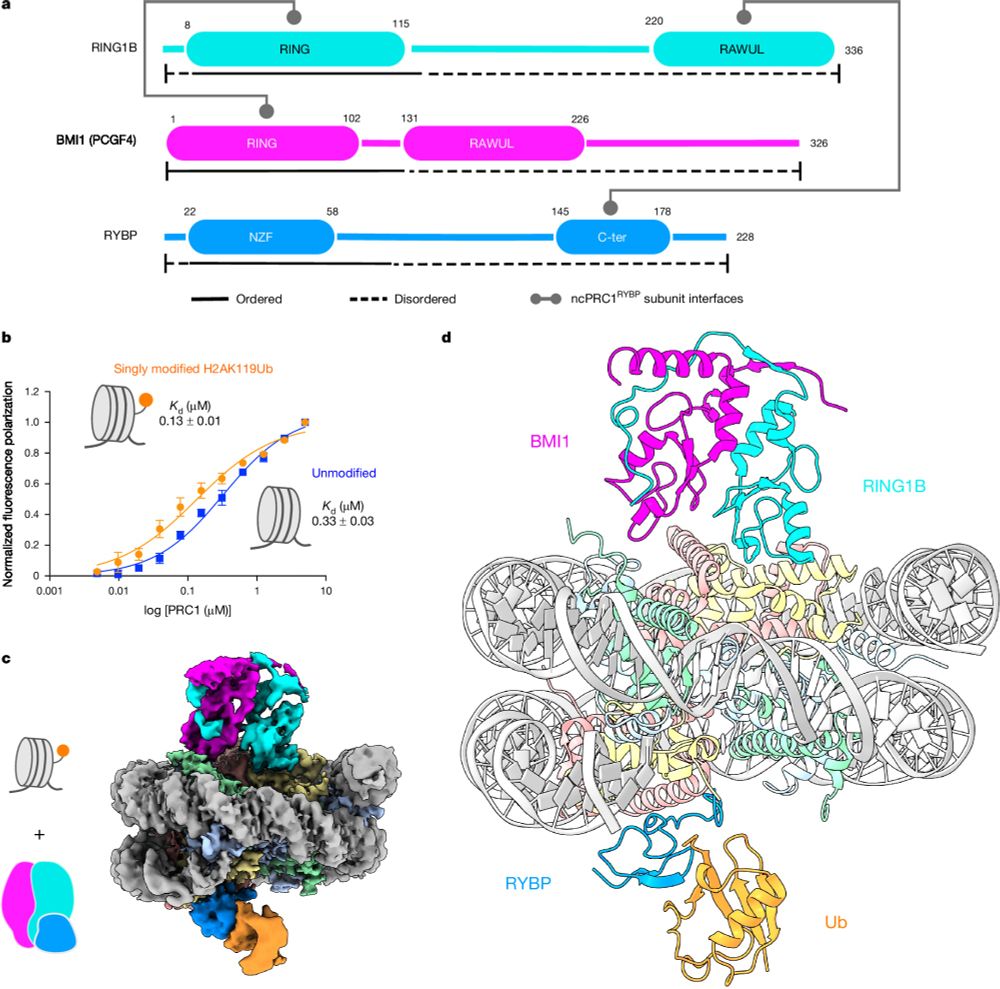
Read–write mechanisms of H2A ubiquitination by Polycomb repressive complex 1
Nature - Cryo-electron microscopy and biochemical studies elucidate the read–write mechanisms of non-canonical PRC1-containing RYBP in histone H2A lysine 119 monoubiquitination and their...
Please see our paper in Nature on read-write mechanisms of H2AK119 ubiquitination by Polycomb repressive complex I. Congrats to the whole team, especially Victoria and huge thanks to our collaborator JP Armache! Also big thanks to Mark Foundation for Cancer Research for the support! rdcu.be/dZ5HZ
13.11.2024 18:23 — 👍 115 🔁 31 💬 7 📌 4
EMDB Team Leader - EMBL-EBI. wwPDB Principal Investigator. Fathering, mixology & DIY enthusiast. #CryoEM #CryoET #firstgen. All opinions are my own.
Biochemist fascinated by visualizing biological processes with the finest spatiotemporal detail; Understand the human immune system to guide novel approaches for combating viral infections and cancer; Postdoc at Cissé Lab / MPI-IE
New PI @mpi_ie, interested epigenetics, development, homologous chromosome interactions and their function (she/her)
Researcher @Jiri Lukas lab, Novo Nordisk Center for Protein Research, Genome Instability, CRISPR-Cas9, heritable consequences of DNA repair, single cell high content screening, greenify research 🌱
Ahh well-a everybody's heard
about the NuRD.
#NuRDistheWord
Cycling/Chromatin Remodelling/Transcription/Enhancers/StemCells
We've got it all.
Living Systems Institute, University of Exeter
https://lsi.exeter.ac.uk/groups/hendrich-group/
Mol bio | Genomics | Development | AI
PhD @Radboud uni, postdoc @ EMBL, Genomics scientist @Google DeepMind
Fragile Nucleosome co-organiser
Views are my own
Study genetic conflicts professionally. Try to avoid conflicts in personal life (with mixed results).
Fred Hutch Basic Sciences,
UW Genome Sciences,
HHMI.
Posting in a personal capacity. My posts don’t reflect my employers’ opinions.
Structure solving and systems learning. Running @solutecarriers. Vienna. Views = my own
PhD student in Narlikar/Ramani labs at UCSF ✨
Professor of Molecular Biology
Head of Department of Cancer Biology and Pathology
University College London
Senior scientist @Princess Margaret Cancer Centre, Prof @UofT. BHAG: conquer cancer by treating it as a disease of the chromatin. https://lupienlab.uhnresearch.ca/
PhD candidate in the Kvon Lab @ UC Irvine 👩🏻🔬🧬
Exploring Hox genes regulation in Stem Cell-Based Embryo Models at @college-de-france.fr | Duboule Lab | CDSN PhD Fellowship
Bioinformatics support for next generation sequencing (NGS) projects with specialisation for MNase-Seq data
Associate Professor & Docent @umeauniversitet.bsky.social
Wallenberg Fellow in Molecular Medicine at WCMM Umeå
Postdoc @embl.org
PhD @cniostopcancer.bsky.social
3D genomics , epigenomics, gene regulation, enhancer, cancer, glioblastoma, cancer neuroscience
We are the Ting Wu Lab @ HMS. Our laboratory studies how chromosome behavior and positioning influence genome function, with implications for gene regulation, genome stability, and disease.
Managed by WuLab members
https://www.transvection.org/home
Studying cellular #stress responses, particularly #senescence and its impact on #immune response, #ageing and #cancer, #tumorigenesis at Cancer Research UK CI @cruk-ci.bsky.social, University of Cambridge @cam.ac.uk
Website: naritalab.com
Group leader at the Centre for Regenerative Medicine (IRR), University of Edinburgh. Exploring the epigenetic mechanisms the control neurodevelopment. Scientist, Dad and lover of all things outdoors!
Assistant Professor @ Stanford University | cancer epigenetics | DNA replication | protein synthesis | vanrechemlab.com
Pain-ter, designer, art tabler✨🌈🌱🇵🇭
Living in Minnesota. they/them
🎨 Portfolio: gawki.net
🎁 Shop is closed: gawki.etsy.com
👋 Next: Planet Comicon
🔴 Stream: www.twitch.tv/gawkiart
‘-> Tuesdays and Fridays @ 6-9 PM CST