
Nature research paper: Semantic design of functional de novo genes from a genomic language model
go.nature.com/48uEnAn
@adititm.bsky.social
BioE PhD student @ Stanford in the Hie Lab // ML for Synthetic Biology

Nature research paper: Semantic design of functional de novo genes from a genomic language model
go.nature.com/48uEnAn

Today in @nature.com, in work led by @adititm.bsky.social, we report the ability to prompt Evo to generate functional de novo genes.
You shall know a gene by the company it keeps!

Published today in @nature.com, @adititm.bsky.social & researchers from the @brianhie.bsky.social lab report that the large-scale genomic model, Evo, is capable of using surrounding genomic context to produce novel, functional genes, enabling an an emergent approach they've termed 'semantic design'.
19.11.2025 18:17 — 👍 16 🔁 8 💬 1 📌 0
This was all possible because of the support of my incredible PI @brianhie.bsky.social and my amazing labmates @samuelhking.bsky.social and Eric Nguyen. Grateful to be surrounded by people who inspire me to be a better scientist.
To learn more, check out the paper: www.nature.com/articles/s41...
Together, this work suggests that genomic sequence models can meaningfully generalize beyond characterized natural evolution. Looking forward, we hope that semantic design can serve as a starting point for function-guided design and optimization of genes across biology.
19.11.2025 16:37 — 👍 2 🔁 0 💬 1 📌 0
Beyond providing novel sequences for functions of interest, SynGenome can be used to predict the roles of domains of unknown function, reveal functional associations across prokaryotic biology, and catalog chimeric proteins with unique domain combinations generated by Evo.
19.11.2025 16:37 — 👍 1 🔁 0 💬 1 📌 0
Semantic design achieved high experimental success rates (up to 50%) without structural conditioning or fine-tuning. To explore semantic design more broadly, we created SynGenome, a database of generations from millions of prompts spanning 9 thousand functional terms.
evodesign.org/syngenome/

Next, we designed anti-CRISPR (Acr) proteins. Evo generated functional Acr proteins that protected against spCas9, despite some having no sequence or predicted structural similarity to known Acrs. This further supported the idea Evo could generalize based on context alone.
19.11.2025 16:35 — 👍 1 🔁 0 💬 1 📌 0
We next asked if semantic design could co-design more evolutionarily diverse sequences. Focusing on toxin–antitoxin systems, we successfully generated a functional RNA antitoxin, a de novo toxic gene, and broadly neutralizing antitoxins. Many had <30% sequence identity to nature.
19.11.2025 16:35 — 👍 1 🔁 0 💬 1 📌 0
We first tested if Evo understands genomic context. Given partial sequences of conserved genes, we show that Evo can achieve near-perfect amino acid sequence recovery and complete entire operons bidirectionally, all while still producing diverse underlying DNA sequences.
19.11.2025 16:34 — 👍 2 🔁 0 💬 1 📌 0
Genomic language models like Evo can leverage this: by prompting with natural genomic context containing genes related to a function of interest, we can ‘autocomplete’ sequences with novel, diverse generations enriched for similar functions. We call this semantic design.
19.11.2025 16:33 — 👍 1 🔁 0 💬 1 📌 0
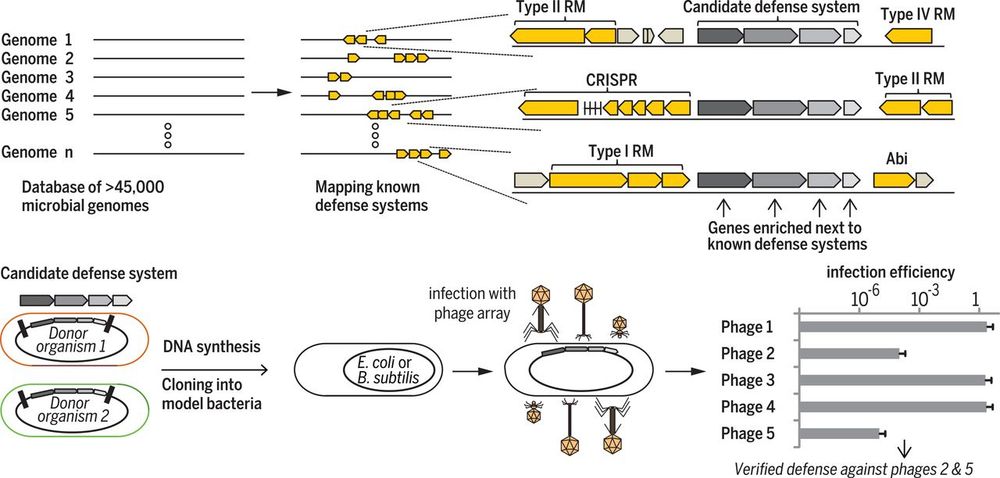
Just as word meaning emerges from context—"you shall know a word by the company it keeps"—prokaryotic gene function is tied to genomic context. This notion of guilt by association, where related genes cluster in operons, has led to the discovery of many molecular tools like CRISPR, BGCs, and more.
19.11.2025 16:33 — 👍 2 🔁 0 💬 1 📌 0
In recent years, we’ve seen immense progress in leveraging generative AI to accelerate biological design. However, using these models to produce diverse sequences with desired high-level functions remains challenging.
19.11.2025 16:32 — 👍 3 🔁 0 💬 1 📌 0What if we could autocomplete DNA based on function?
Today in @Nature, we share semantic design—a strategy for function-guided design with genomic language models that leverages genomic context to create de novo genes and systems with desired functions. 🧵
www.nature.com/articles/s41...
New @biorxiv-synthbio.bsky.social on #Evo 👀⤵️🧵 introducing Evo 1.5 for semantic mining + SynGenome - an AI-generated genomics database #AI #synbio #LLM🧬 @adititm.bsky.social @brianhie.bsky.social et al. @arcinstitute.org
22.12.2024 19:23 — 👍 11 🔁 3 💬 3 📌 0If you’re interested in learning more or have any questions or feedback, definitely reach out! The preprint, along with a link to the PDF (since bioRxiv seems to be having some server issues) are linked below! N/N
www.biorxiv.org/content/10.1...
evodesign.org/Semantic_Min...
This work was a massive collaborative effort between my amazing fellow graduate students Samuel King and Eric Nguyen! And of course, none of this would have happened without the incredible mentorship of @brianhie.bsky.social! Very fortunate to work with such inspiring scientists daily :) 13/N
19.12.2024 18:54 — 👍 1 🔁 0 💬 1 📌 0Ultimately, this study suggests that biological sequence models may be able to nontrivially generalize beyond known evolutionary space and that prompt engineering can be a valuable tool for steering generation towards desired functional outcomes. 12/N
19.12.2024 18:54 — 👍 0 🔁 0 💬 1 📌 0
SynGenome is publicly available at evodesign.org/syngenome/. You could use SynGenome to find diversified natural proteins, functionally characterize uncharacterized genes, or find highly divergent proteins with potentially conserved functions. We’re excited to see what the community can find! 11/N
19.12.2024 18:54 — 👍 0 🔁 0 💬 1 📌 0To generate SynGenome, we used prompts derived from the genes encoding prokaryotic proteins in UniProt, reasoning that the resultant generations may be enriched for functions related to the proteins the prompts were derived from. 10/N
19.12.2024 18:54 — 👍 1 🔁 0 💬 1 📌 0
Finally, to apply semantic mining to generate functional genes from across prokaryotic biology, we developed SynGenome, a database containing over 120 billion base pairs of synthetic DNA sequences. 9/N
19.12.2024 18:54 — 👍 1 🔁 0 💬 1 📌 0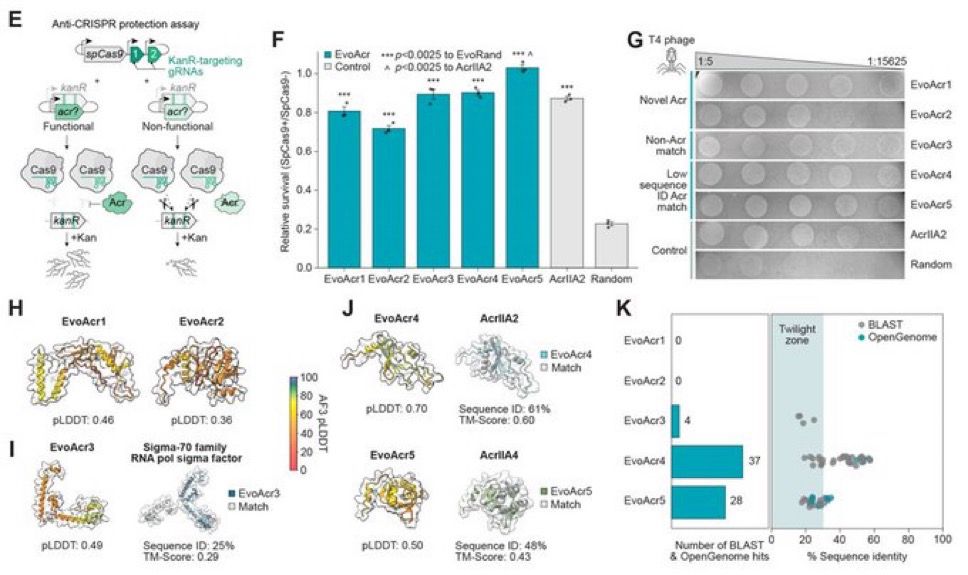
Despite this high diversity, 17% of the Acr designs we tested were functional. Additionally, many of our experimentally validated Acrs had low confidence AF3 structure predictions and two eluded significant structural or sequence characterization, making them akin to “de novo” genes (!) 8/N
19.12.2024 18:54 — 👍 1 🔁 0 💬 1 📌 0
We then applied semantic mining to see if we could design new anti-CRISPR (Acr) proteins, a highly diverse group of proteins with limited sequence or structural conservation thought to sometimes emerge via de novo gene birth. 7/N
19.12.2024 18:54 — 👍 2 🔁 0 💬 1 📌 0
Half of the Evo-designed antitoxins we experimentally tested were functional (!), with most possessing only remote homology to natural proteins and some appearing to neutralize diverse toxin classes. 6/N
19.12.2024 18:54 — 👍 1 🔁 0 💬 1 📌 0
We then applied semantic mining to generate a multi-gene bacterial toxin-antitoxin (TA) system. Using context from known TA systems as prompts, we first designed and experimentally validated a toxin gene. This toxin gene then served as a prompt for Evo to generate new conjugate antitoxins. 5/N
19.12.2024 18:54 — 👍 1 🔁 0 💬 1 📌 0
As an initial test, we first demonstrated that Evo 1.5, a new version of Evo with extended pretraining, was able to understand genomic context, showing that it could complete highly conserved genes and operons when prompted with only partial sequences. 4/N
19.12.2024 18:54 — 👍 1 🔁 0 💬 1 📌 0
Taking inspiration from genome mining techniques using guilt-by-association, we hypothesized that by prompting Evo with a gene encoding a desired function, we could guide the model to generate a new gene with a related function. We term this approach “semantic mining.” 3/N
19.12.2024 18:54 — 👍 1 🔁 0 💬 1 📌 0Just as words derive meaning from their context, DNA gains functional significance within the context of genes, operons, and genomes. In prokaryotes, genes with related functions are often grouped together in close proximity on the DNA sequence. 2/N
19.12.2024 18:54 — 👍 1 🔁 0 💬 1 📌 0
Excited to have the first project of my PhD out!! By leveraging genomic language model Evo’s ability to learn relationships across genes (i.e., "know a gene by the company it keeps"), we show that we can use prompt-engineering to generate highly divergent proteins with retained functionality. 🧵1/N
19.12.2024 18:54 — 👍 19 🔁 5 💬 1 📌 1