New preprint 🚨 We systematically measured 17 million phospho-specific dose-response curves (133 kinase inhibitors × 5 cell lines) to decrypt the kinases that shape the human phosphoproteome. We show that drug perturbation potency (not effect size) links kinases to substrates while controlling FDR.
20.11.2025 11:59 — 👍 22 🔁 7 💬 0 📌 0

Hello Toronto!🍁The Terrific TUM Team is happy to be attending #HUPO2025. Kusterlab, Wilhelmlab and Leelab have a diverse set of presentations for you that we can't wait to share 🤓.
We're excited to spend the next days reuniting with old friends and making new connections. See you there!
09.11.2025 11:33 — 👍 17 🔁 6 💬 0 📌 1

Thanks @tum.de for highlighting Jakob's recent paper in @cellpress.bsky.social in the university news:
www.ls.tum.de/en/ls/public...
If you haven't had the chance to read the publication, you can check it out here: doi.org/10.1016/j.ce...
07.08.2025 12:26 — 👍 13 🔁 2 💬 0 📌 0
What makes it even better: All experiments were performed in a dose-resolved manner—so you can dive into dose-response curves in our interactive dashboards! #CurveCurator #BeautifulCurves (3/4)
31.07.2025 08:10 — 👍 0 🔁 0 💬 1 📌 0
We explore how inhibition of oncogenic KRAS signaling reshapes protein phosphorylation, ubiquitylation, and protein expression—across multiple time points, using various KRAS inhibitors, as well as upstream and downstream pathway inhibitors, in different KRAS-mutant cell lines. (2/4)
31.07.2025 08:10 — 👍 0 🔁 0 💬 1 📌 0
Kusterlab is looking for a new member to join our Bioinformatics team.
Apply now if you're interested in working with us!
30.07.2025 12:56 — 👍 2 🔁 1 💬 0 📌 0

Another day of #ASMS2025 in Baltimore, and we're back with another poster! Make sure to visit Flo today - he'll present the latest insights from his large-scale decryptM project.
Go #TeamMassSpec!
04.06.2025 09:28 — 👍 6 🔁 1 💬 0 📌 1

Hello Baltimore! We're looking forward to #ASMS2025 and all the new developments in mass-spec and proteomics.
If you're attending today, make sure to check out @msleelab.bsky.social's poster on citrullination in multiple sclerosis. Sophia 'Lapo' Laposchan is excited to see you there!
02.06.2025 08:14 — 👍 4 🔁 1 💬 0 📌 0
New paper!
Our chemoproteomic survey of phenylhydroxamic acids identifies the first drug-like inhibitors for nucleotide-binding protein HINT1 and nucleoside kinases NME1-4. On top, we found probes for the HDAC inhibitor off-target MBLAC2. Check out the open-access article! tinyurl.com/yh92vh6b
09.05.2025 12:41 — 👍 6 🔁 1 💬 0 📌 0

3/2025 Issue ➡️ www.embopress.org/toc/17444292...
metabolic mutations drive bacterial antibiotic resistance evolution, prior knowledge for GRN inference, HPA drugs fail mood disorder clinical trials
Cover: ATR kinase inhibitors work together to overcome chemoresistance @kusterlab.bsky.social
12.03.2025 09:55 — 👍 8 🔁 2 💬 0 📌 0
🔎 To uncover the molecular mechanisms driving this synergy, we employed both chemo- and phosphoproteomics (decryptM), which revealed how ATR inhibition disrupts the DNA damage response when combined with gemcitabine. (3/4)
04.03.2025 16:06 — 👍 2 🔁 1 💬 1 📌 0
🧫 Using large-scale phenotypic drug screening, we identified promising drug combinations that enhance tumor cell death - and found Gemcitabine and ATR inhibitors to synergize most effectively. (2/4)
04.03.2025 16:06 — 👍 0 🔁 0 💬 1 📌 0
We'll DM you
04.02.2025 05:10 — 👍 0 🔁 0 💬 0 📌 0

Our tool was built using Vue.js and D3.js and is free for public reuse (github.com/kusterlab/bi..., github.com/wilhelm-lab/...).
Thanks to everyone involved! 🥳 We're looking forward to seeing how it'll be received by the community. (6/6).
10.01.2025 14:20 — 👍 2 🔁 0 💬 0 📌 0
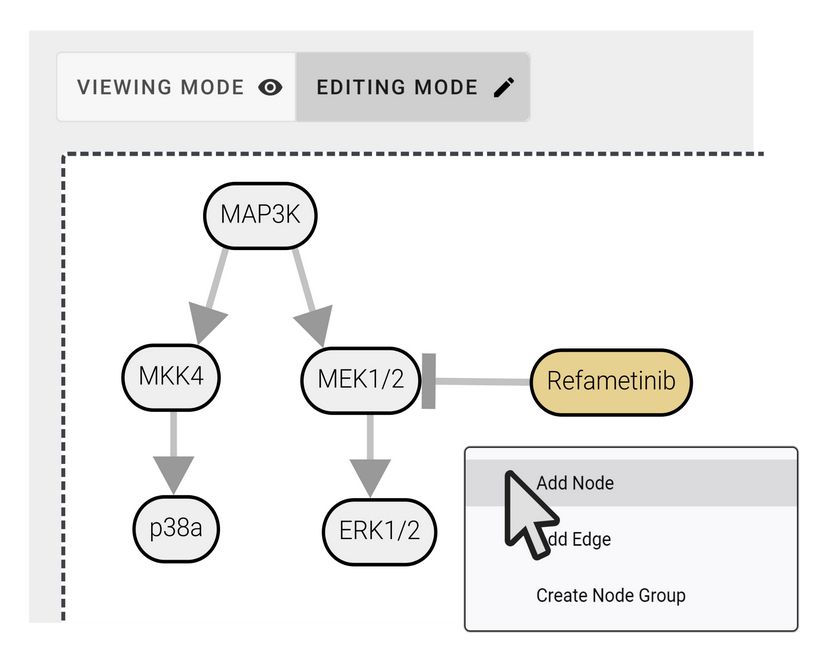
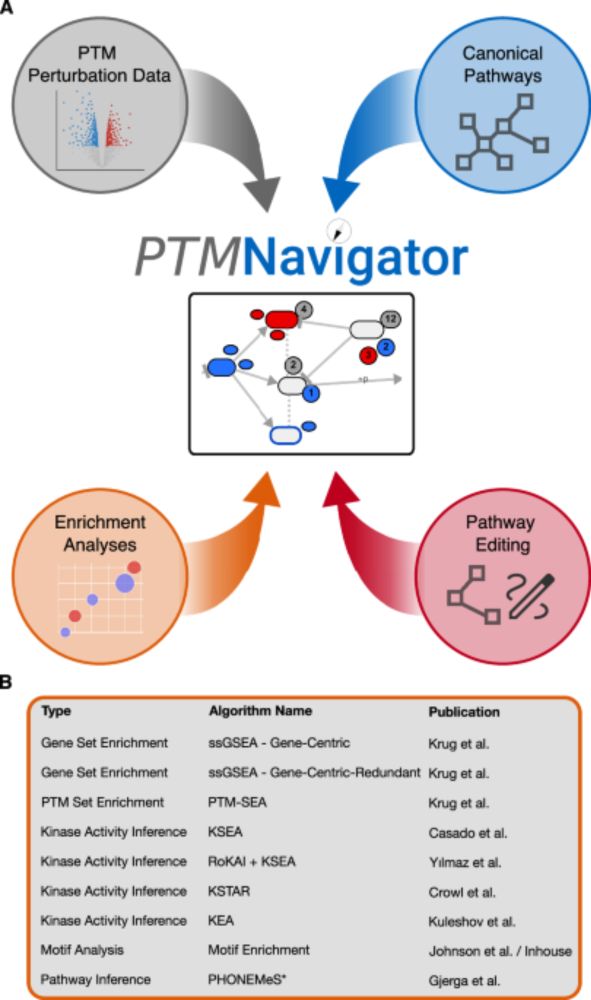
If canonical pathway diagrams are not enough for you, just build your own! PTMNavigator's editing mode allows you to design custom pathways, tailored to your data and research question. 🧑🔬 (5/6)
10.01.2025 14:20 — 👍 2 🔁 0 💬 1 📌 0
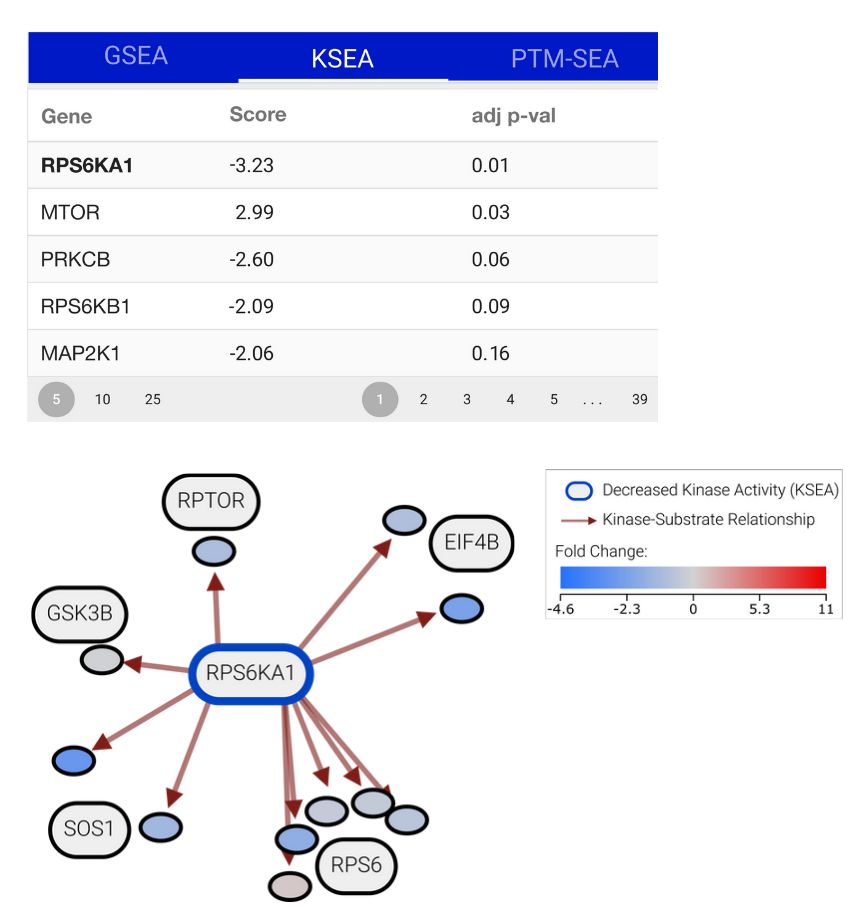
What's more, we developed a server that runs various enrichment analyses on your data (github.com/kusterlab/en...) 🤓. The results of this are automatically integrated into PTMNavigator and help you figure out which pathways, proteins and PTM signatures are potentially relevant for your dataset.(4/6)
10.01.2025 14:20 — 👍 1 🔁 0 💬 1 📌 0
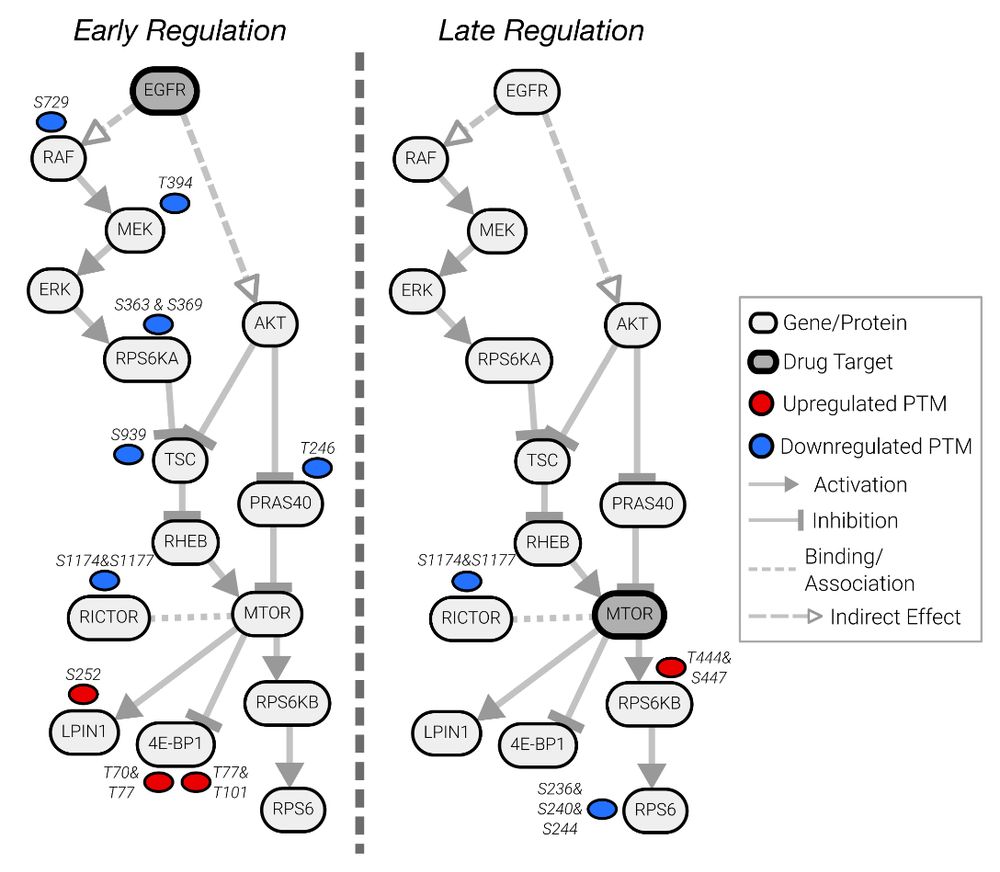
PTMNavigator projects your data onto interactive pathway diagrams, showing you where the hotspots of PTM regulation are. 🗺️ 📍 This can be used, for example, to contrast how two drugs affect the same pathway at different stages. (3/6)
10.01.2025 14:20 — 👍 1 🔁 0 💬 1 📌 0
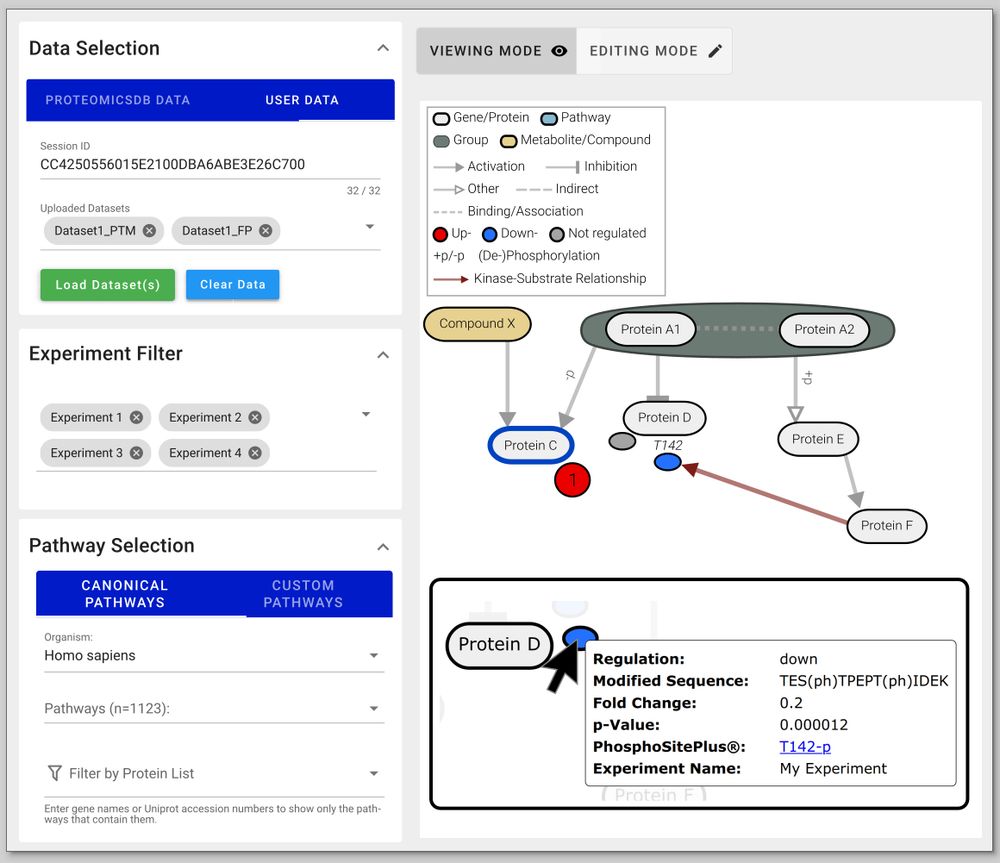
Putting PTM data into context can be a daunting task, and it's easy to lose sight of the big picture. 👀
To tackle this, Julian Müller and colleagues have developed PTMNavigator, which is available at ProteomicsDB 💻 www.proteomicsdb.org/analytics/ptmNavigator (2/6)
10.01.2025 14:20 — 👍 1 🔁 0 💬 1 📌 0
Postdoc @ Küster lab | prev, PhD @ Trost lab | proteomics | chemoproteomics | mass spectrometry | target deconvolution | drug MoA
PhD student in proteomics & mass spectrometry @kusterlab
Bioinformatics group leader @kusterlab.bsky.social
Visit my personal website: thematthew.me
Proteomics and metabolomics | Bacterial proteomics | opinions shared here are my own
Cutting-edge research, news, commentary, and visuals from the Science family of journals. https://www.science.org
Head of the Core Facility for Omics Sciences at IRCCS Ist. G. Gaslini - Pediatric Hospital, interested in mass spectrometry applied to life sciences. Opinions are my own.
Life scientist.
Head of EMBO Membership & Elections and Courses & Workshops @embo.org #EMBOevents
previously: Editor at Molecular Systems Biology, EMBO Press
Views my own
data scientist @ TUM || proteomics, human microbiome, and clinical health research
The German Society for Proteome Research (DGPF) unites academia and industry to advance proteomics. It fosters innovation, collaboration, and technology transfer while supporting young talents - the next generation of proteomics researchers.
We are the Savitski lab located @ EMBL using and developing proteomics methods for assessing the state of the proteome
A research group interested in proteomics, peptidomics, and PTMomics in neuro-diseases📍TU_Muenchen
| CLINSPECT-M consortium | opinions are my own (Chien-Yun)
#OpenAccess @embopress.org journal publishing pioneering research in systems & synthetic biology, systems medicine
Induced Proximity, Epigenetics, Drug Discovery, Proteomics.
EMBO/MSCA postdoctoral fellow in Winter lab @ CeMM/AITHYRA
Focus on developing proteomics technologies.
https://www.chem.ox.ac.uk/people/shabaz-mohammed
https://www.bioch.ox.ac.uk/research/mohammed
https://scholar.google.co.uk/citations?hl=en&user=6FdXeiwAAAAJ&view_op=list_works&sortby=pubdate
PostDoc in Olsen Group and Arachnid wll-being executive manager. 🕷️🕸️ Working on Spatial Proteomics and extracellular vesicles. 🤗
PhD student @Kusterlab working on protein-RNA interactions
Active Mirrored Profile: https://bsky.app/profile/maxischuh.mastodon.social.ap.brid.gy