
Excited to share our work on inhibitors turning degraders now out: doi.org/10.1038/s415...
27.11.2025 06:13 — 👍 30 🔁 6 💬 1 📌 1@sevlechner.bsky.social
Induced Proximity, Epigenetics, Drug Discovery, Proteomics. EMBO/MSCA postdoctoral fellow in Winter lab @ CeMM/AITHYRA

Excited to share our work on inhibitors turning degraders now out: doi.org/10.1038/s415...
27.11.2025 06:13 — 👍 30 🔁 6 💬 1 📌 1
🧬Kinase inhibitors that degrade?
New study by CeMM, @aithyra.bsky.social & @irbbarcelona.org reveals that kinase inhibitors can also trigger the degradation of their target proteins, becoming a new tool for drug development & #TDP research.
➡️ More: bit.ly/3LY6jor
📄 Paper: bit.ly/43MffU1
New preprint 🚨 We systematically measured 17 million phospho-specific dose-response curves (133 kinase inhibitors × 5 cell lines) to decrypt the kinases that shape the human phosphoproteome. We show that drug perturbation potency (not effect size) links kinases to substrates while controlling FDR.
20.11.2025 11:59 — 👍 22 🔁 7 💬 0 📌 0
Did you ever come across a phosphosite in your proteomics data for which nothing was known? - I bet so!
We have developed a new strategy termed "potency coherence analysis" that leverages the drug potency dimension in decryptM to decode the kinases that shape the human phosphoproteome.
Read more:

New preprint: We isolate peptide–RNA photo-crosslinks with tunable RNA chains from living cells for mass spec. This maps over 4,700 crosslinking sites across 744 proteins and offers the first glimpse into the RNA sequences in crosslinks by MS. Read here: doi.org/10.1101/2025...
09.09.2025 11:46 — 👍 24 🔁 14 💬 0 📌 1
Our new review paper about Pathway-Centric PTM Data Analysis is out this week in #Proteomics!
We cover databases, enrichment tools, software for pathway reconstruction, and full-fledged platforms that help to interpret high-throughput PTM datasets.
Check it out here: doi.org/10.1002/pmic...

They DecryptED all the KRAS small molecule inhibitors! Including cysteine enrichments! proteomicsnews.blogspot.com/2025/08/decr...
13.08.2025 13:17 — 👍 10 🔁 3 💬 2 📌 0Measuring & modeling nucleic acids is very useful and needed.
Yet, it provides a partial view.
It misses many biological processes, from signal transduction and metabolic fluxes, through enzyme activities to therapeutic effects.
bsky.app/profile/slav...

@aithyra.bsky.social Opening Symposium "AI for Life Science" with Nobel Laureate Frances Arnold as keynote speaker in addition to an outstanding line up of speakers on a variety of topics across biological scales and data modalities.
Registration is now open at cemm.at/aithyra-symp...
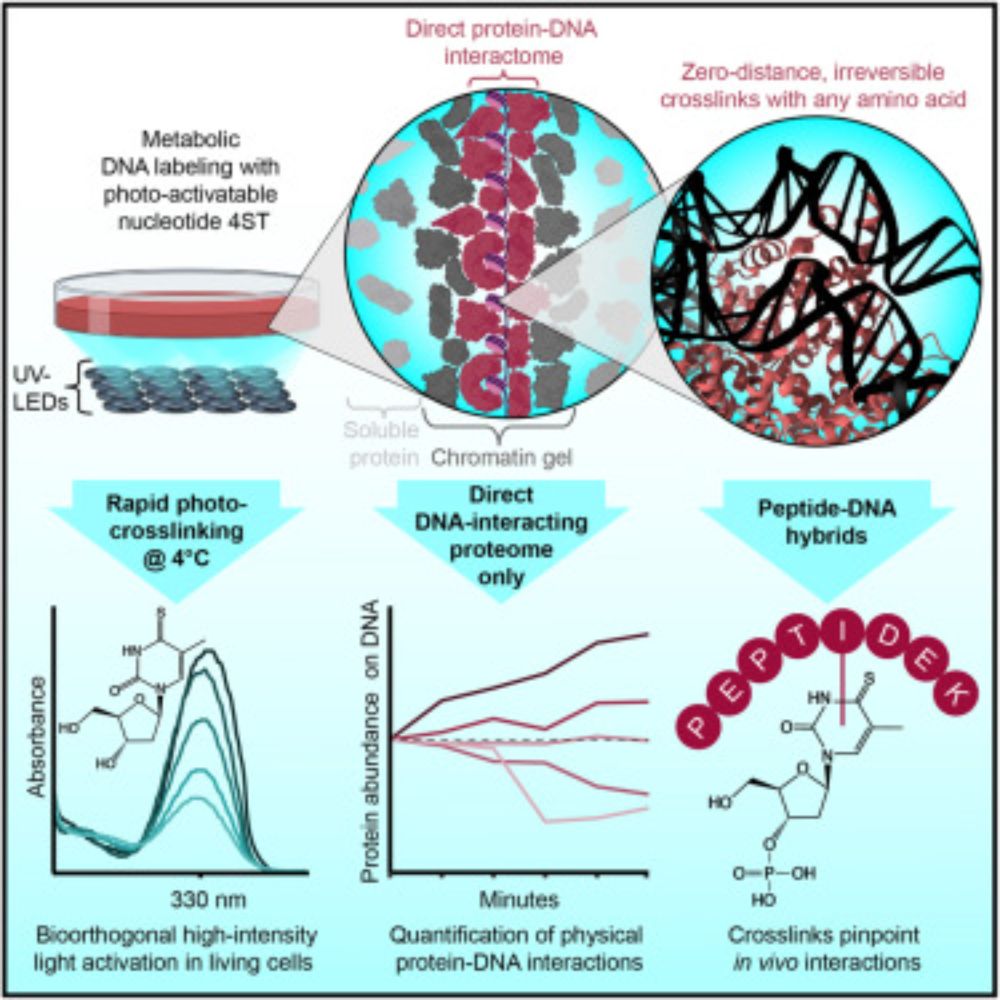
🚨Our new paper is online🚨
We use zero-distance⚡photo-crosslinking⚡to reveal direct protein-DNA interactions in living cells, enabling quantitative analysis of the DNA-interacting proteome on a timescale of minutes. #DNA #Chromatin #Proteomics
www.cell.com/cell/fulltex...
Key takeaways:
a) every second HDAC inhibitor with a phenylhydroxamic warhead inhibits MBLAC2
b) NME1-4 and HINT1 are now targetable with small molecule inhibitors
c) chemoproteomics is always good for surprises 🙂
We characterized HINT1 and NME1-4 inhibition with enzyme activity assays, molecular docking, and co-crystal structures.
09.05.2025 12:00 — 👍 0 🔁 0 💬 1 📌 0As often seen with chemoproteomic approaches, we made some unexpected but exciting findings: several molecules bound to the yet undrugged proteins NME1-4 and HINT1.
09.05.2025 12:00 — 👍 0 🔁 0 💬 1 📌 0Having profiled over 100 molecules, we derive rules for which HDAC pharmacophores are prone to inhibit MBLAC2. We also found nanomolar MBLAC2 inhibitors with >30-100-fold selectivity over HDACs.
09.05.2025 12:00 — 👍 0 🔁 0 💬 1 📌 0Here, we profiled a hand-picked library of metalloenzyme inhibitors. This uncovered several unreported off-targets of HDAC inhibitors and confirmed MBLAC2 as a frequent off-target, also for purportedly selective inhibitors such as SW-100.
09.05.2025 12:00 — 👍 0 🔁 0 💬 1 📌 0
Using a chemoproteomic competition assay, we previously identified MBLAC2 as off-target of more than 20/50 profiled HDAC inhibitors (www.nature.com/articles/s41...)
09.05.2025 12:00 — 👍 0 🔁 0 💬 1 📌 0Paper alert!
We report the first inhibitors for nucleoside kinases NME1-4 and for the nucleotide-binding protein HINT1.
On top, we provide probes for MBLAC2, an off-target of every second (!) hydroxamic acid-based HDAC inhibitor!
tinyurl.com/yh92vh6b
👇🧵
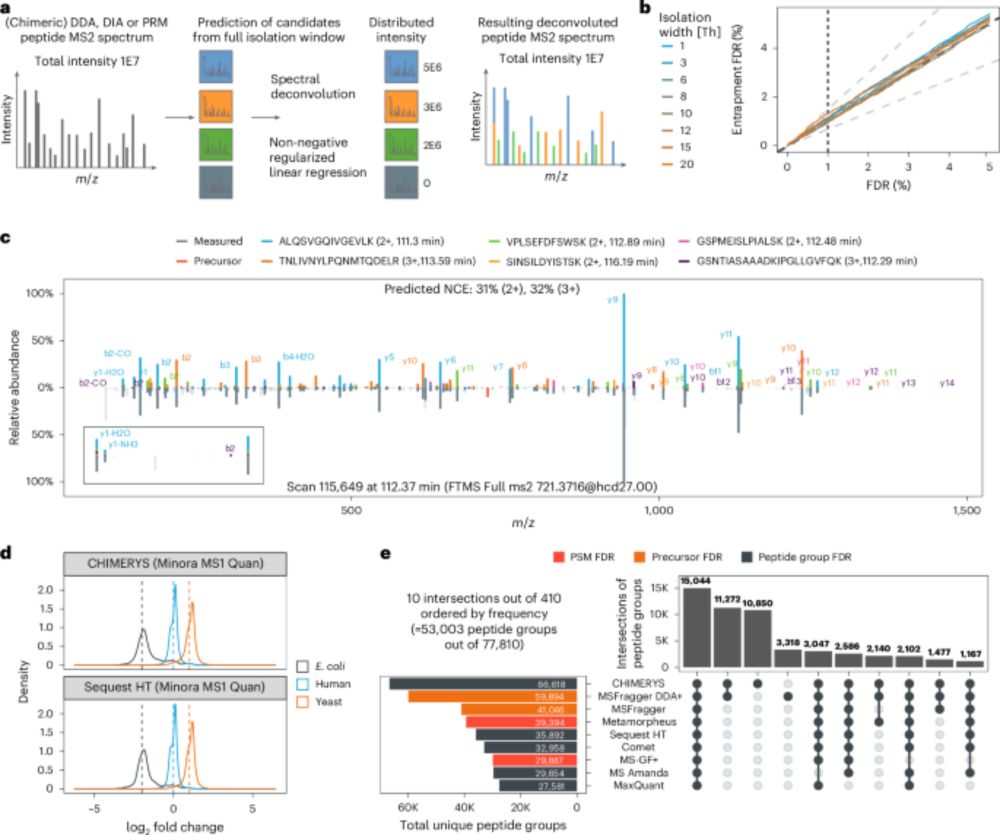
One algorithm to rule them all? CHIMERYS bridges the gap — DDA,DIA and PRM — together at last!
With #CHIMERYS, we can now directly compare DDA and DIA data — 🍎 to 🍎 finally made possible.
doi.org/10.1038/s41592-025-02663-w
#KusterLab #WilhelmLab #MSAID #Proteomics
🚨 Hiring Alert – Senior Scientist (Biology) 🚨
My lab is looking for a senior scientist to lead functional genomics efforts and decode how small molecules reshape biology
🧬 Lead screens
🤝 Collaborate with top-tier AI/chemistry/biology minds
Interested? then apply: cemm.onlyfy.jobs/job/t7lhh1kz

🚨 Rational discovery of VHL molecular glues. New Preprint from @amgen.bsky.social Induced Proximity group led by postdoc Jonathan Bushman reports the discovery of a VHL molecular glue degrader of GEMIN3 by Picowell RNA-seq. Read on for details. 🧵1/8 www.biorxiv.org/content/10.1...
21.03.2025 04:17 — 👍 32 🔁 6 💬 1 📌 2
Less than 3 weeks left⏳Register by April 7 for the #Ubiquitin & Friends Symposium, May 8-9, in beautiful Vienna!
Fantastic guest speakers, open slots for short talks, flash talks& posters: Great opportunity for students & #ECRs! Register soon to save your spot➡️ www.protein-degradation.org/symposium/
Did you recently complete a ChemBio/MedChem PhD (non-UK) and are looking for the next challenge? I am seeking a prospective applicant for a Newton Fellowship with an exciting project in targeted protein dephosphorylation! Please reach out to e.devita@qmul.ac.uk
royalsociety.org/grants/newto...
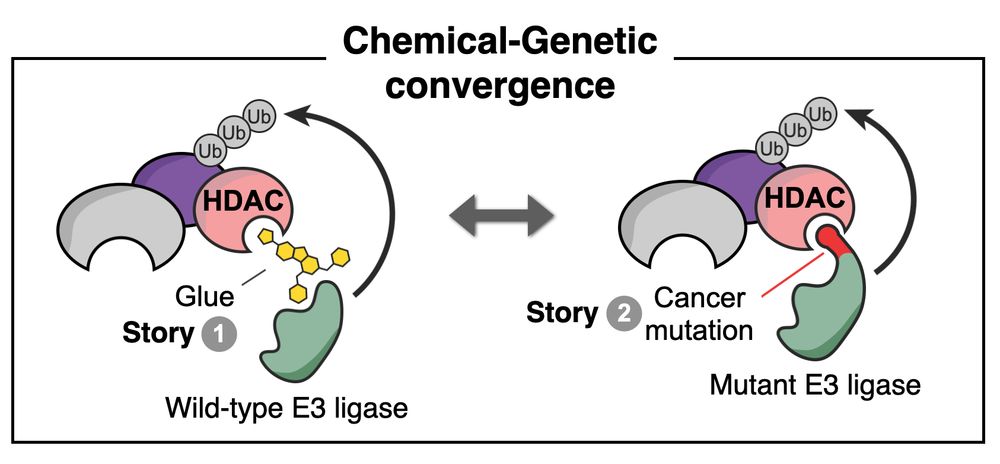
Today in @nature.com we share our back-to-back stories with Ning Zheng’s lab revealing chemical-genetic convergence between a molecular glue degrader & E3 ligase cancer mutations. 1/5
12.02.2025 16:20 — 👍 88 🔁 40 💬 5 📌 5