HT-PELSA, a new proteomics tool by EMBL researchers, processes samples 100x faster and works directly with complex crude cell, tissue, and bacterial lysates – developments which could accelerate drug discovery and basic biological research 💊
🔗 www.embl.org/news/science...
05.11.2025 10:09 — 👍 28 🔁 10 💬 2 📌 0
Happy to see our HT-PELSA paper now published in @natsmb.nature.com 🎊 Big thanks for the constructive review process! 📖Read the manuscript here (www.nature.com/articles/s41...) & check the thread for additional information ⬇️
05.11.2025 10:15 — 👍 36 🔁 14 💬 0 📌 1

So grateful and honored to receive the MCP Lectureship Award at the #ASBMBProteomics meeting at the Broad Institute! Huge thanks to the wonderful organizers and to everyone—past and present—who’s been part of our lab.
20.08.2025 10:09 — 👍 42 🔁 7 💬 2 📌 0
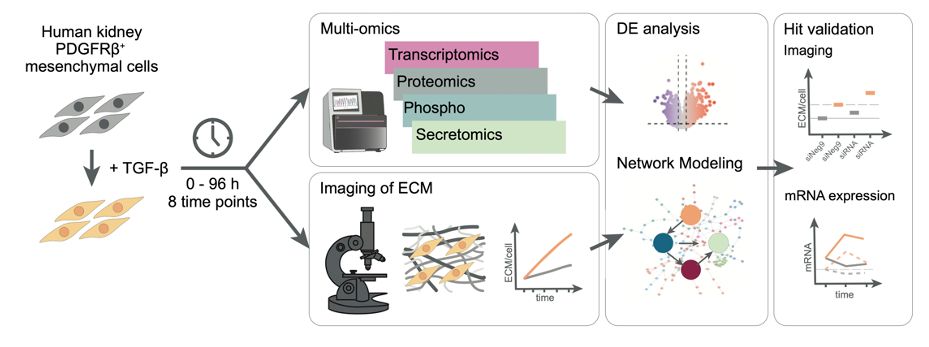
The final version of our multi-omics study on kidney fibrosis is out now (tinyurl.com/kidneyfibMSB). Together w/ Pepperkok + Savitski labs @embl.org, we present a time-resolved #multiomics + computational network modeling approach in combination w/ phenotypic assays to study #kidneyfibrosis
27.06.2025 13:29 — 👍 22 🔁 11 💬 1 📌 1
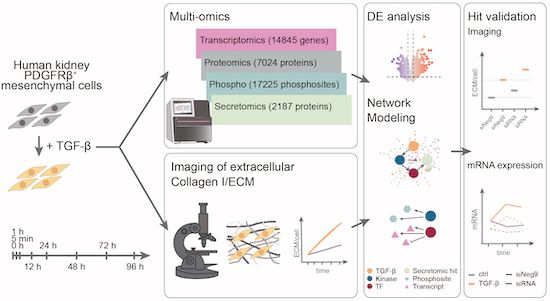
integrated temporally resolved transcriptomic, proteomic, and secretomic profiling of human kidney mesenchymal cells from @savitski-lab.bsky.social @saezlab.bsky.social @miraburtscher.bsky.social identify dynamic changes upon TGF-beta stimulation ➡️ www.embopress.org/doi/full/10....
03.07.2025 12:30 — 👍 4 🔁 3 💬 0 📌 0
Thank you for this great perspective on our #glycoproteomics study @timveth.bsky.social and @nmriley.bsky.social!
09.05.2025 14:49 — 👍 14 🔁 1 💬 0 📌 0
PELSA APP
Data can be explored 2u7b8b-nico0h0ttmann.shinyapps.io/PELSAAPP/;
This work was led by Kejia Li @kejiali.bsky.social and Clément Potel with contributions from Isabelle Becher, Nico Hüttmann @nicohuettmann.bsky.social , Martín Garrido-Rodríguez
@martingarridorc.bsky.social, and Jennifer Schwarz.
30.04.2025 07:56 — 👍 4 🔁 0 💬 0 📌 0
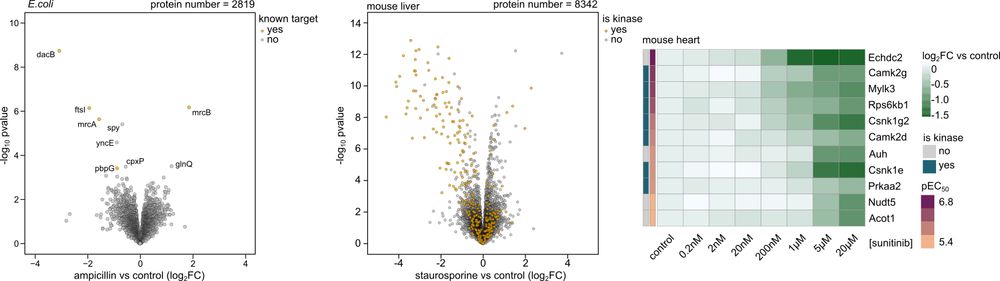
3/3 We extended its application to crude human cell line, bacteria, and tissue lysates and sensitively identify membrane targets! It works extremely well in tissues which allowed us to identify off-targets for sunitinib in mouse heart tissue and could explain its well-known cardiotoxicity.
30.04.2025 07:51 — 👍 4 🔁 0 💬 1 📌 0
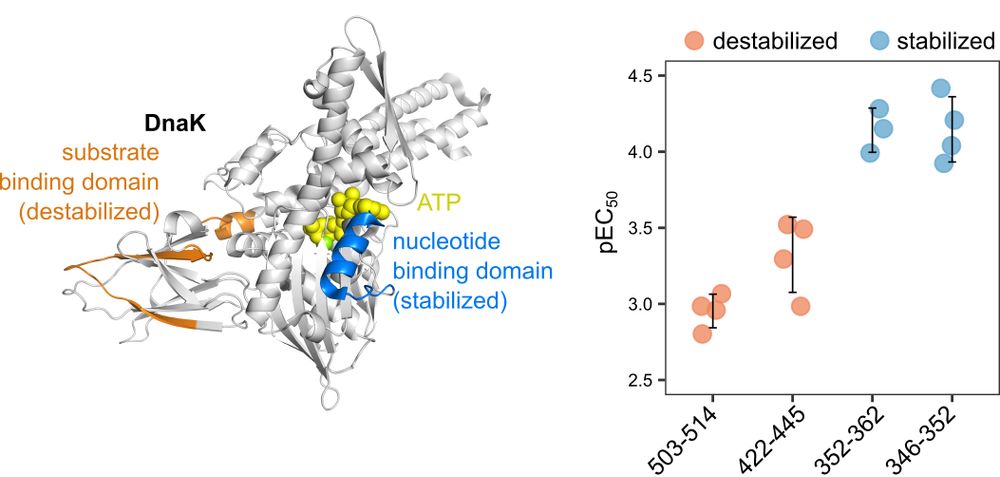
2/3 We provide a large collection of ATP-binding proteins in E.coli and give rich information on how they respond to ATP. Our data nicely showed how chaperon protein DnaK binds ATP at ATP-binding region at low ATP concentration and dissociates the substrate protein at high ATP concentration.
30.04.2025 07:50 — 👍 3 🔁 0 💬 1 📌 0
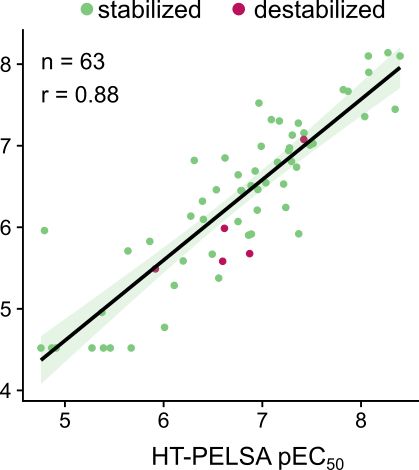
1/3 We extended the original PELSA protocol to work with 96-well plates and show a 100-fold higher throughput, excellent reproducibility, accurate determination of drug-binding affinities in dose-response experiments as shown by a strong correlation of 0.88 with kinobeads-based measurements.
30.04.2025 07:48 — 👍 3 🔁 0 💬 1 📌 0
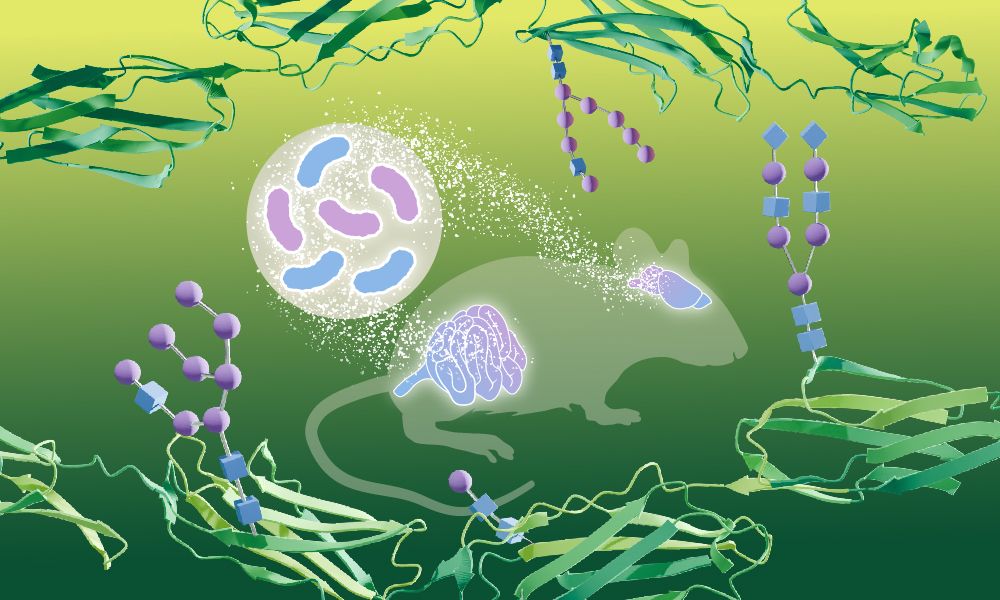
Bacteria can influence how sugars modify proteins in the brain – shown for the first time by EMBL researchers.
In their study, the scientists describe a new method to study glycosylation systematically & quantitatively, leading to new biological insights. 🧪🧠📈
www.embl.org/news/science...
10.02.2025 10:17 — 👍 46 🔁 13 💬 0 📌 0
DQGlyco explorer
All data can be interactively explored 🖥️ apps.embl.de/glycoapp/. This work was led by Clement Potel, @miraburtscher.bsky.social and @martingarridorc.bsky.social. We thank our great collaborators @zimmermannlab.bsky.social and @typaslab.bsky.social.
10.02.2025 10:20 — 👍 2 🔁 0 💬 0 📌 0
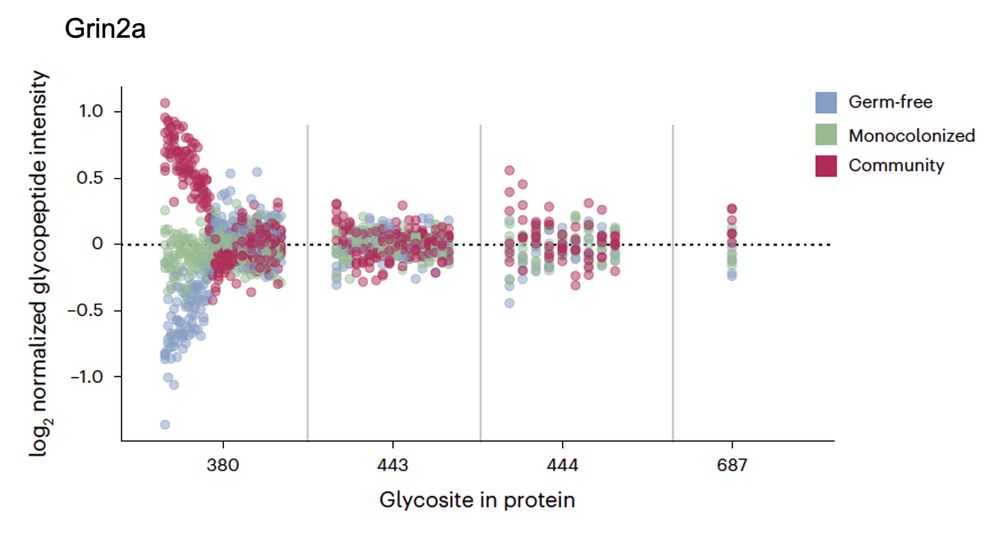
Finally, we once again uncovered site-specific modulation of glycosylation upon perturbation, meaning that only some glycoforms on some glycosites are modulated, even within the same protein, suggesting complex regulatory mechanisms.
10.02.2025 10:20 — 👍 1 🔁 1 💬 1 📌 0
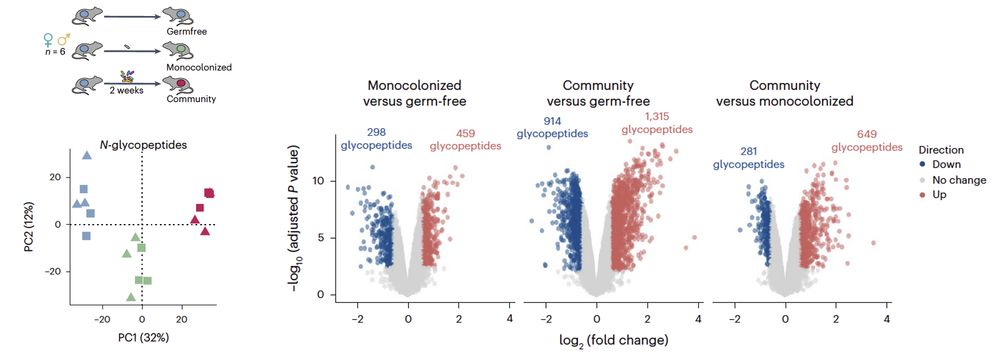
Next, we showed remodeling of the mouse brain glycoproteome upon gut microbiome colonization 🐭 The link b/w gut microbiome & brain physiology is long known, but molecular mechanisms remain elusive. We showed that proteins involved in neurotransmission & axon guidance were particularly affected.
10.02.2025 10:20 — 👍 2 🔁 0 💬 1 📌 0
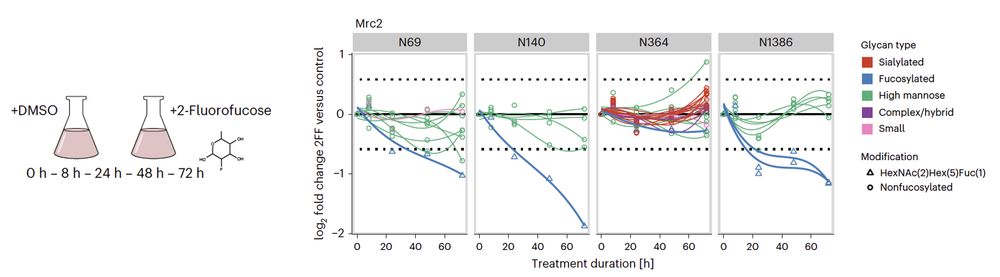
We next used our quantitative approach to measure the dynamics of glycosylation changes in human cells treated with a fucosylation inhibitor 🧫 We discovered pervasive site-specific modulation of glycosylation upon perturbation.
10.02.2025 10:20 — 👍 1 🔁 1 💬 1 📌 0
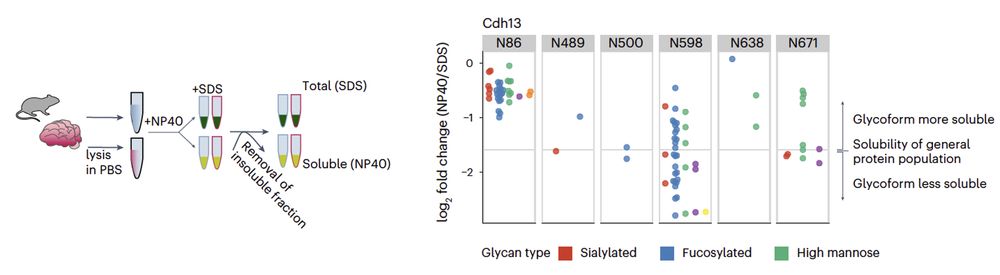
Very little is known about glycoform functionality. We developed a functional glycoproteomics approach enabling the proteome-wide characterization of the solubility of different glycosylated proteoforms in the mouse brain 🐭
10.02.2025 10:20 — 👍 3 🔁 0 💬 1 📌 0
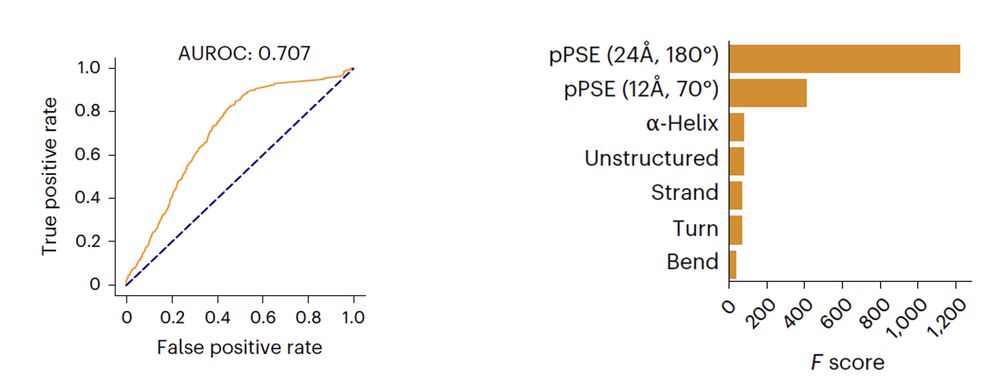
To understand structural features governing the level of site microheterogeneity, we leveraged our comprehensive dataset in combination with AlphaFold DB 🖥️
10.02.2025 10:20 — 👍 3 🔁 1 💬 1 📌 0
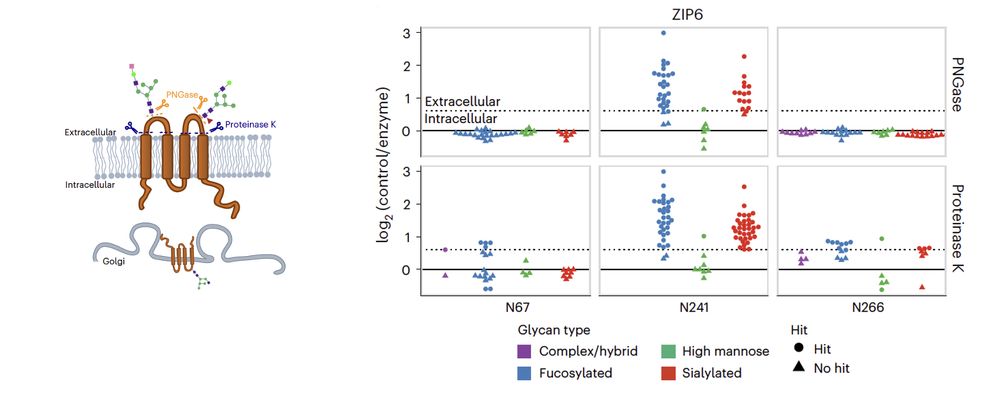
It is essential to differentiate surface-exposed glycoforms from intermediate species. To address this, we treated intact living human cells with two enzymes, enabling proteome-wide characterization of mature glycoforms 🧫
10.02.2025 10:20 — 👍 2 🔁 0 💬 1 📌 0
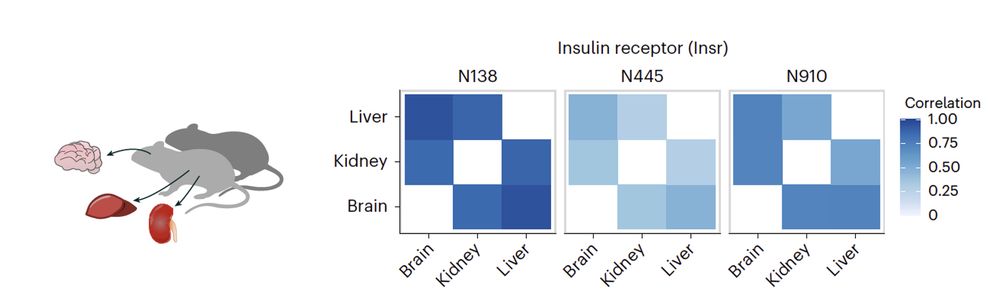
Using our quant approach, we discovered that many sites have similar glycosylation patterns across tissues 🐭However, we identified a subset of sites driving tissue-specificity, enriched in domains involved in cell adhesion, signaling & immunity. E.g., only N445 exhibits tissue specificity on INSR.
10.02.2025 10:20 — 👍 2 🔁 0 💬 1 📌 0
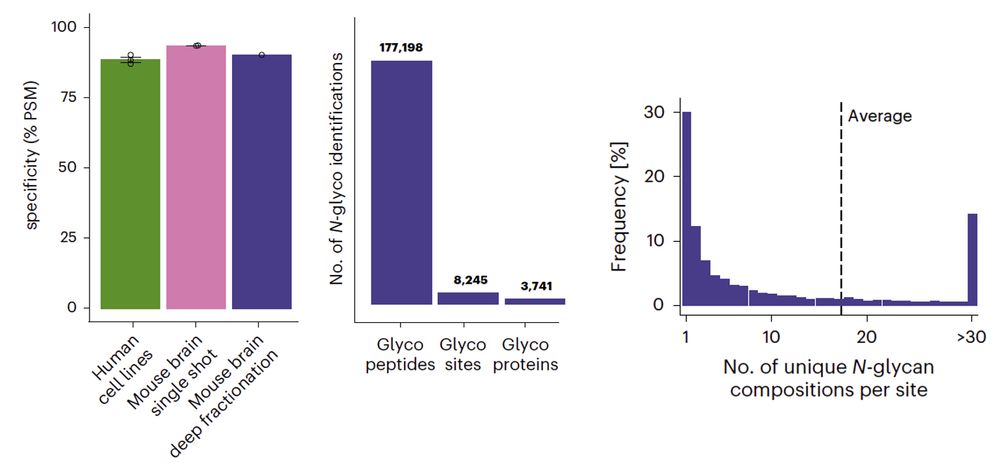
We are very happy to present our work on N-glycoproteomics!🍬 Our method enables the selective enrichment and precise quantification of intact N-glycopeptides to explore the dynamics of glycosylation microheterogeneity.
www.nature.com/articles/s41...
10.02.2025 10:20 — 👍 40 🔁 19 💬 1 📌 3
Proteins | Sugars | Yeast | Beer | Mass Spectrometry | Professor in Biochemistry UQ. CW chickens, fungi, cats, pups.
Human Technopole, Milan, 6-8 October 2025
Staff Scientist in Mass spectrometry platform @fmp-berlin.de. Interested in cross-linking mass spectrometry and interactomics.
Professor of Molecular Systems Biology, University of Cambridge
Focus on developing proteomics technologies.
https://www.chem.ox.ac.uk/people/shabaz-mohammed
https://www.bioch.ox.ac.uk/research/mohammed
https://scholar.google.co.uk/citations?hl=en&user=6FdXeiwAAAAJ&view_op=list_works&sortby=pubdate
Absea Biotechnology | Empowering protein science
14,000+ recombinant proteins & 11,000+ monoclonal antibodies covering 70% of the human proteome.
Germany-USA-China
https://www.absea.bio/index.html
The Sorek Lab
Weizmann Institute of Science, Israel
https://www.weizmann.ac.il/molgen/Sorek/
PhD student in the Savitski Team @EMBL Heidelberg. Alum Berezovski lab and JLHMS facility @uOttawa, @TUDarmstadt. Handball and Tennis and more coming
Postdoc in Savitski lab, EMBL, Heidelberg. Interested in functional proteomics, microbiology, metabolism, and drug target discovery.
Postdoc Fellow at Saez-Rodriguez and Savitski labs (EMBL). Original from Córdoba, Spain.
Assistant professor at Umeå University and Team leader at the Laboratory for Molecular Infection Medicine Sweden (MIMS)
Follow also lab account: @mateuslab.com
Alumnus at @uu.se and at the @EMBL.org (@savitski-lab.bsky.social and @typaslab.bsky.social)
Our lab at Umeå University uses proteomics to understand protein function and interactions in human gut microbiome species.
Predoc @savitski_lab @EMBL | Computational Biology @saezlab | Interested in functional proteomics and systems biology 🧫🧬
Team Coordinator Proteomics Services, EMBL-EBI.
omics, data science, and coding
Lead of bigbio projects
Group Leader at Charité – Universitätsmedizin Berlin. Proteomics technologies and applications, DIA-NN author. https://aptila.bio
Dynamics of Living Systems (https://www.frohlichlab.com) group leader @thecrick.bsky.social Understanding signalling & cell state dynamics through mathematical modelling and machine learning.
I'm a scientist at Harvard Medical School and I specialize in mass spectrometry, proteomics, and bioinformatics. I spend most of my time thinking about protein-protein interactions!
Assistant Professor at UW Genome Sciences developing/applying proteomic technologies | he/him/his | Views are my own.
Professor of Genome Sciences University of Washington, Seattle. Interested in proteomics and mass spectrometry.