https://lms.mrc.ac.uk/work/vacancies/research-assistant-2/
Great opportunity for someone to join our dynamic group @cvgenomics.bsky.social as a research assistant, where we are using genomics and transcriptomics to better understand heart disease. Closing date 27th August for applications!
@mrc-lms.bsky.social
@jamesware.bsky.social
t.co/u68MZ6DDFD
21.08.2025 07:04 — 👍 1 🔁 1 💬 0 📌 0
Now available in the PGS Catalog 🔗: www.pgscatalog.org/publication/...! Thanks to authors like Sean for submitting the data directly, find out how to submit your data at pgscatalog.org/submit
26.02.2025 14:42 — 👍 7 🔁 3 💬 1 📌 0
See the attached 🧵 for some of our main takeaways
18.02.2025 17:24 — 👍 1 🔁 0 💬 0 📌 0
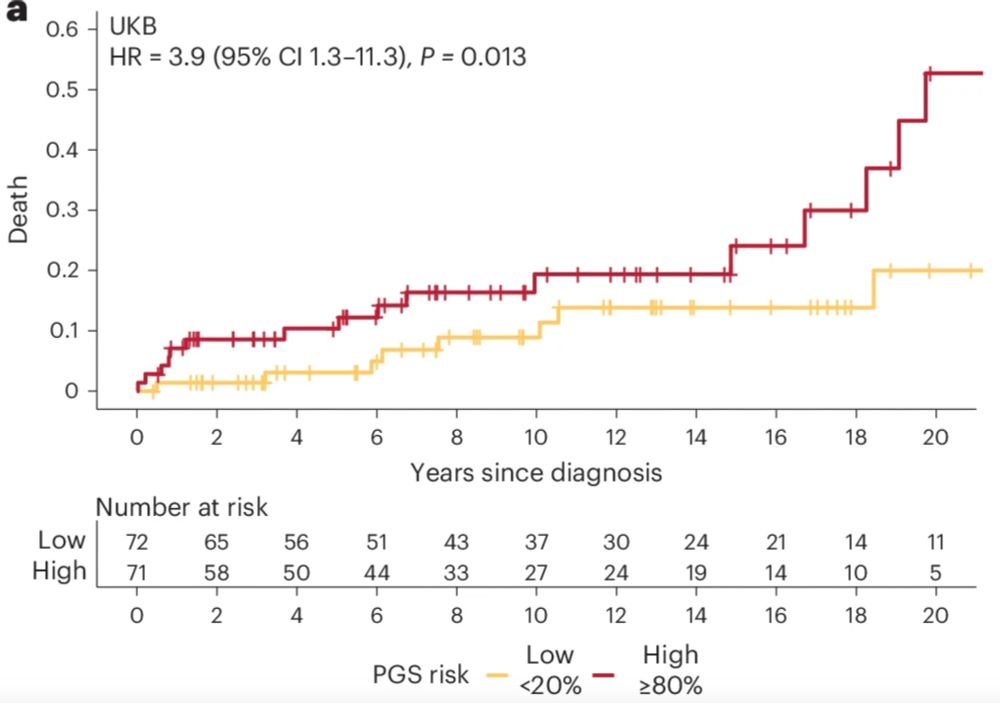
And finally, although many people with HCM lead relatively normal lives, some unfortunately develop CV complications. In >1K people with HCM in UK Biobank and 100K, PGS acts as a novel biomarker stratifying survival and MACE after diagnosis. Future work needed to fit this into clinical models. n/n
18.02.2025 15:29 — 👍 0 🔁 0 💬 0 📌 0
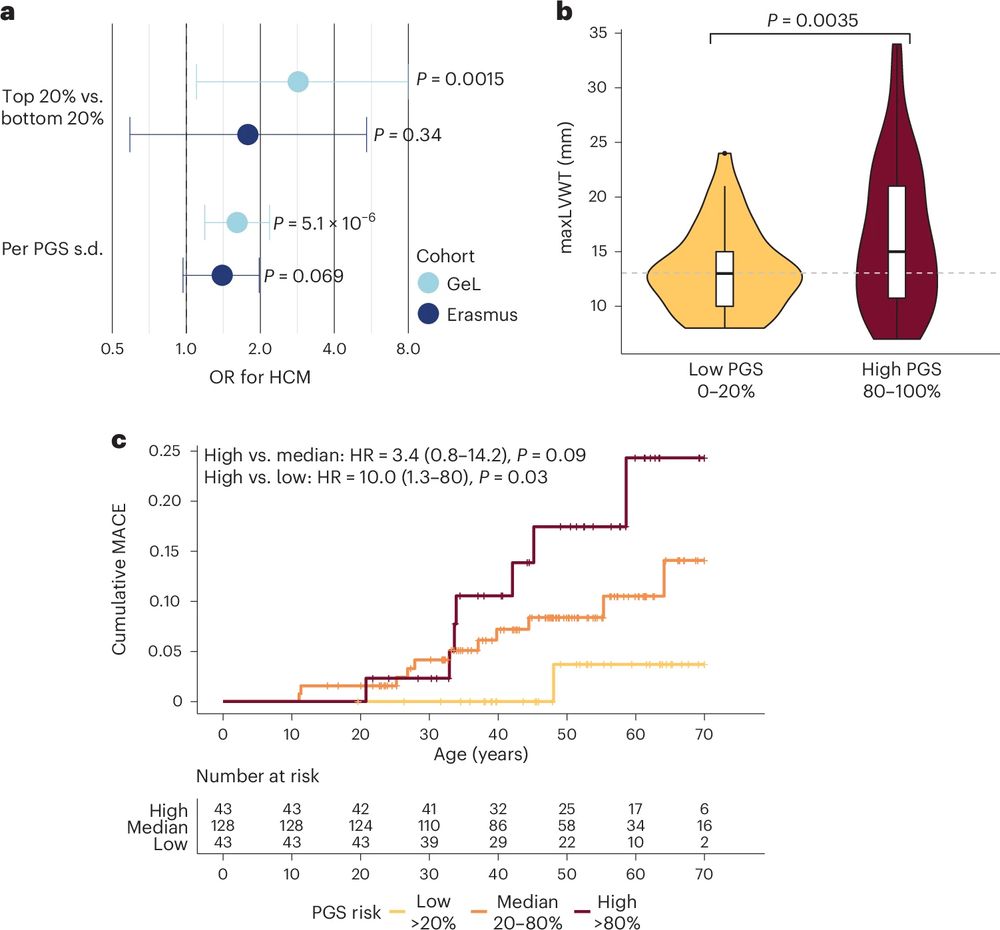
An area of clinical importance is risk assessment in patient’s families. Most relatives who are pheno-ve remain under long-term surveillance. PGS can stratify future HCM risk, HCM severity, and adverse CV outcomes among relatives, providing yet another opportunity for personalised care. 4/n
18.02.2025 15:27 — 👍 0 🔁 0 💬 1 📌 0
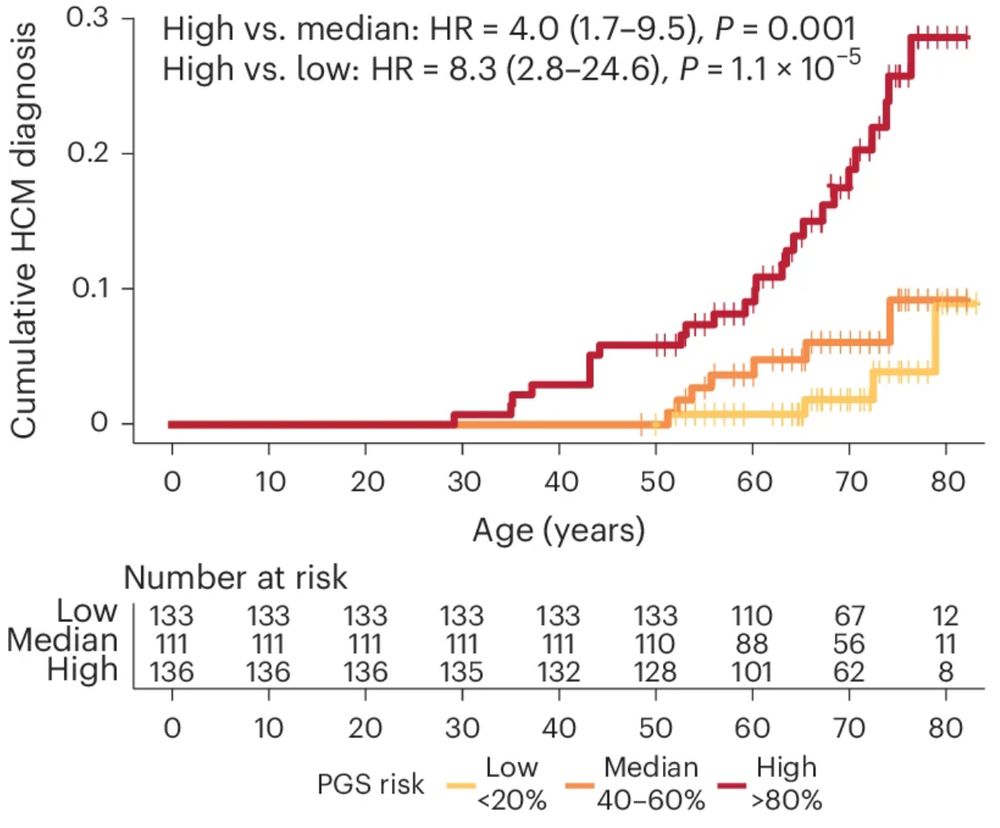
Pathogenic variant carriers in HCM genes don’t all go on to develop HCM (incompletely penetrant). In >1.2K carriers from UKB + 100K, PGS acts as a key modifier of rare variant effects = potential to guide personalised surveillance/early intervention strategies in secondary finding settings. 3/n
18.02.2025 15:26 — 👍 0 🔁 0 💬 1 📌 0
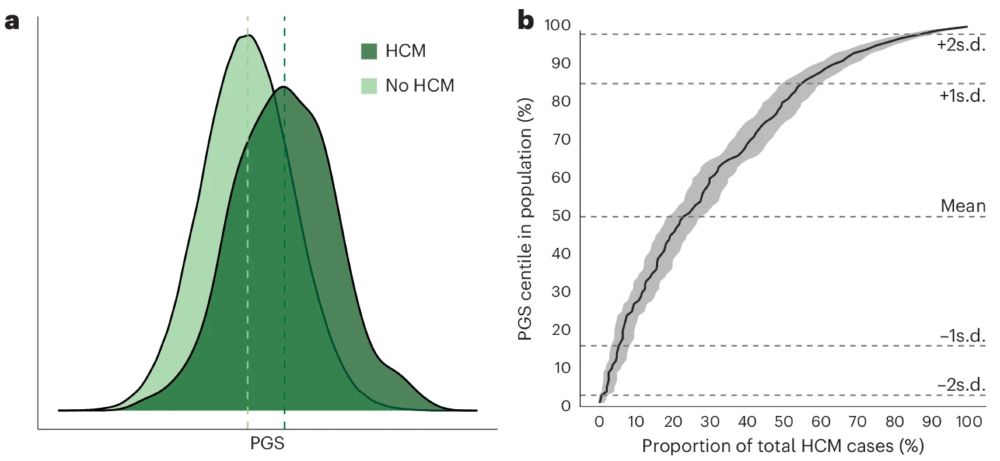
We show in the UK Biobank that PGS associates with HCM, with 15-fold increased risk of HCM for those with scores in top 1% vs. mean. Almost half of all people with HCM have a score >1SD from the mean, ~20% >2sd. 2/n
18.02.2025 15:24 — 👍 0 🔁 0 💬 1 📌 0
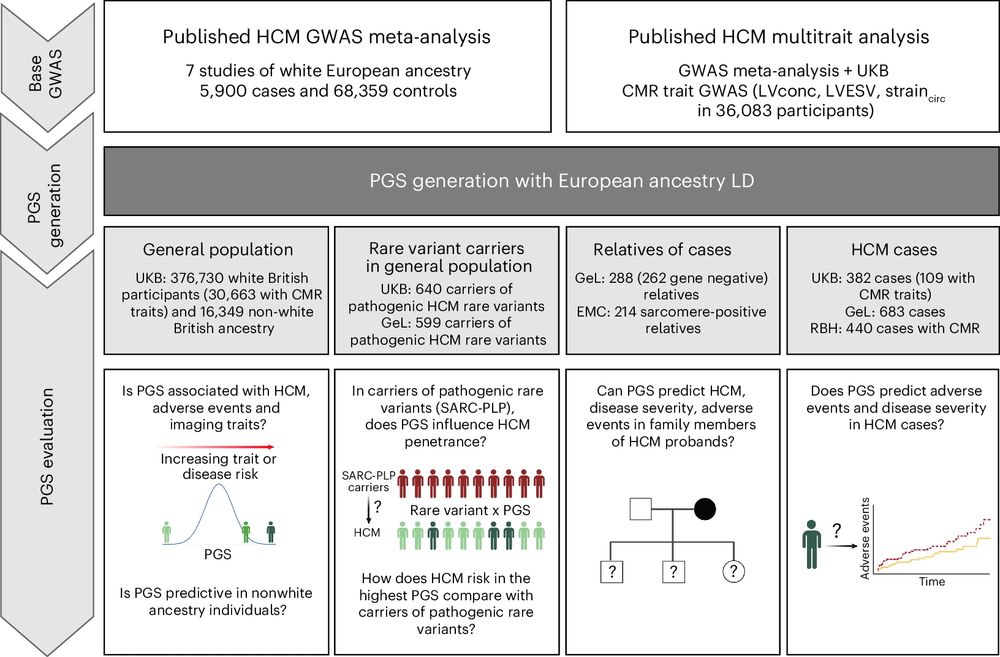
🧵 Polygenic scores in hypertrophic cardiomyopathy
📎 Paper: www.nature.com/articles/s41... in @naturegenet.bsky.social
We created PGS using sumstats from our linked GWAS study www.nature.com/articles/s41..., evaluating it across a range of clinical settings in several cohorts. 1/n
18.02.2025 15:21 — 👍 2 🔁 2 💬 1 📌 1
🚨Our double header on genetics of hypertrophic #cardiomyopathy out today in @naturegenet.bsky.social 🧬
📍GWAS www.nature.com/articles/s41...
📍polygenic scores www.nature.com/articles/s41...
A consortia effort by @jamesware.bsky.social Hugh Watkins @conniebezzina.bsky.social Rafik Tadros & Anuj Goel
18.02.2025 12:19 — 👍 11 🔁 4 💬 0 📌 1
Genome-wide association analyses in Nature Genetics provide insights into the molecular etiology of dilated cardiomyopathy, which may inform the design of genetic testing strategies and may facilitate the development of targeted therapeutics. https://go.nature.com/4fIkNmh
Genome-wide association analyses in Nature Genetics provide insights into the molecular etiology of dilated cardiomyopathy, which may inform the design of genetic testing strategies and may facilitate the development of targeted therapeutics. https://go.nature.com/4fIkNmh 🧪
27.11.2024 19:08 — 👍 52 🔁 10 💬 2 📌 1
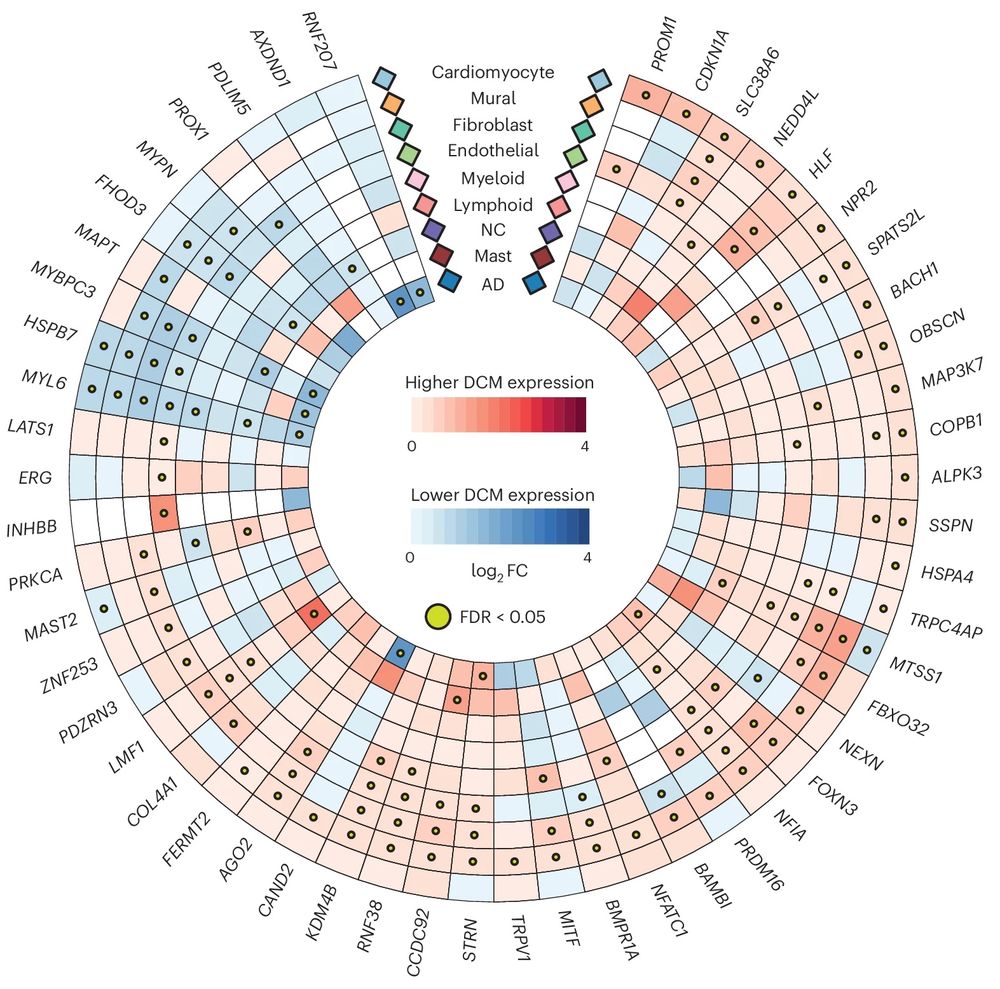
And finally, using snRNA-seq of 52 end-stage heart failure DCM samples, we identify the causal cell types, and explore changes in expression of GWAS effector genes in disease, and identify important intercellular interactions for several of the genes (COL4A1, BMPR1A, etc.)
5/5
21.11.2024 13:53 — 👍 2 🔁 0 💬 0 📌 0
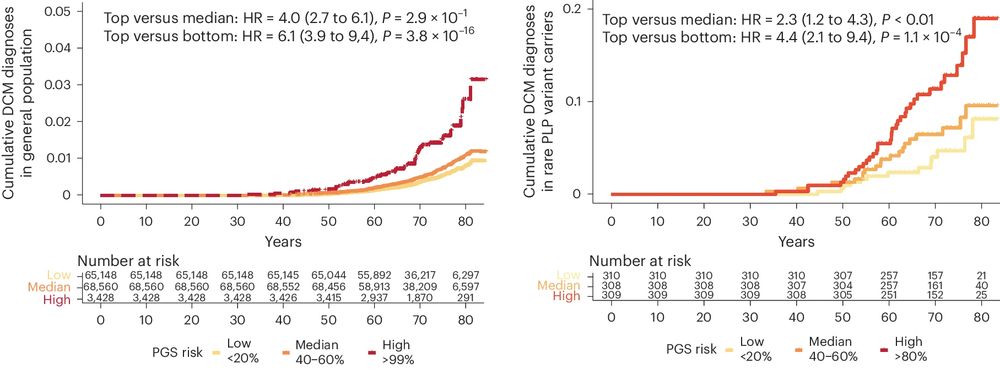
Having highlighted the importance of SNPs in DCM risk in this study, we next created a polygenic risk score and show that it predicts DCM in the population, and in 1,546 carriers of rare DCM-causing variants.
4/5
21.11.2024 13:49 — 👍 1 🔁 0 💬 1 📌 0
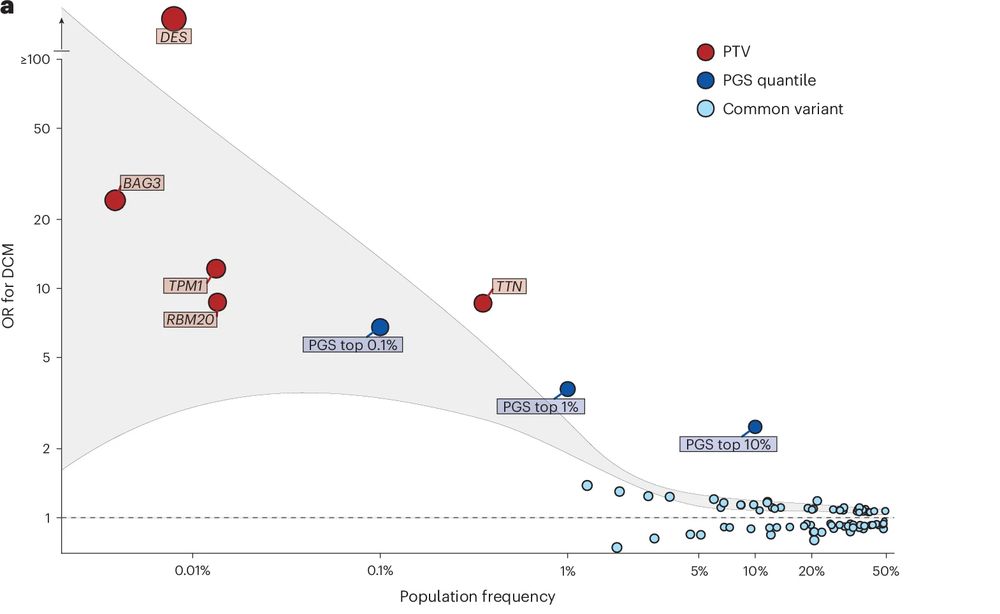
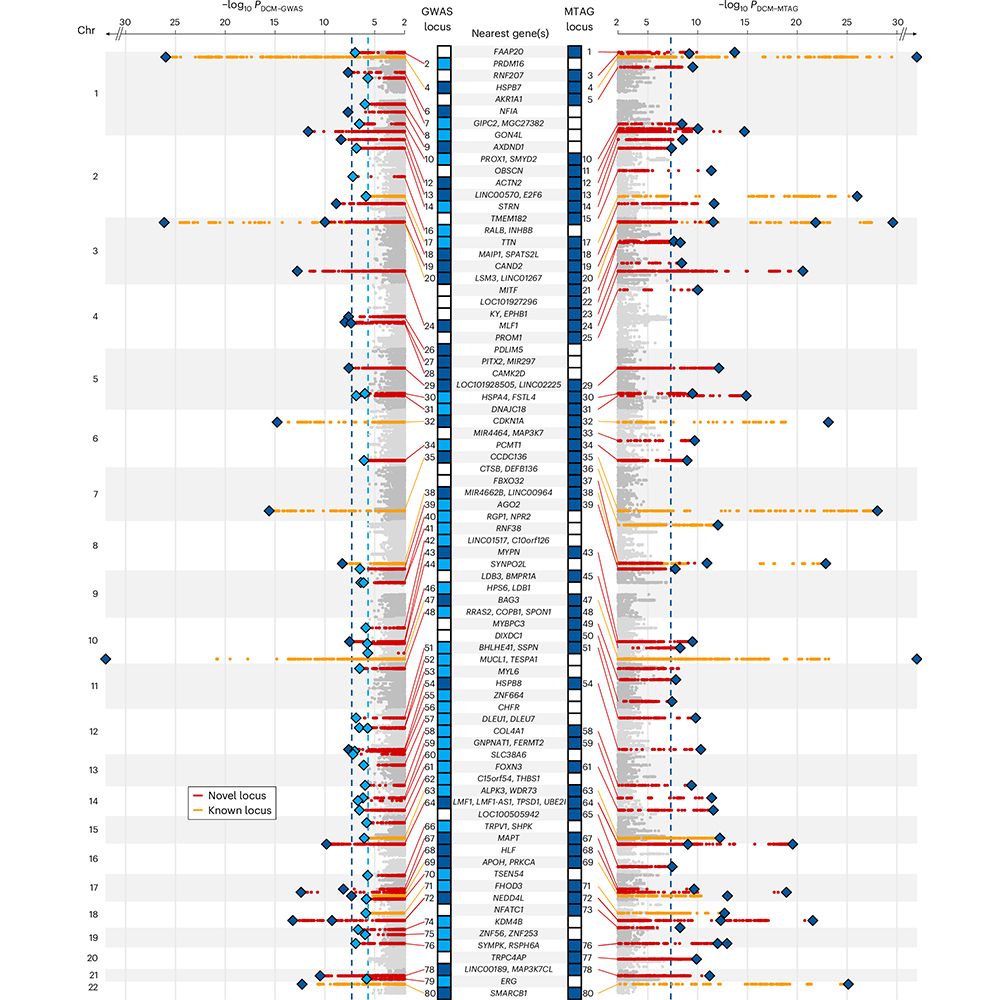
Highlighting overlap of common and rare variant causes of DCM across the spectrum of allele frequencies, 7 genes known to cause DCM were identified. Using UKBB and 100K Genomes Project we also discover 3 rare novel genetic causes of DCM (MAP3K7, SSPN, and NEDD4L).
3/5
21.11.2024 13:47 — 👍 1 🔁 0 💬 1 📌 0
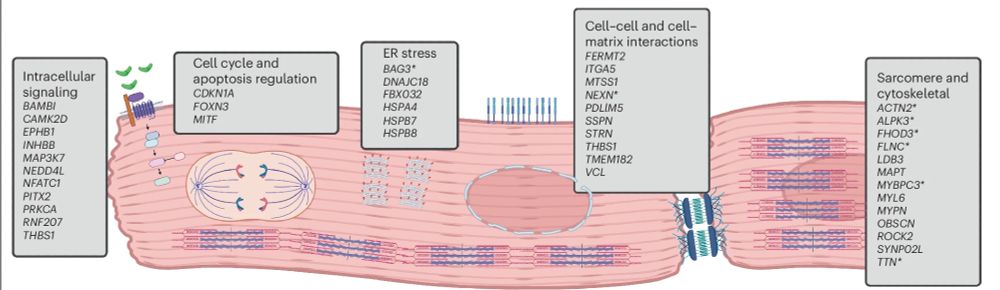
We incorporate 8 in silico tools to identify effector genes throughout the genome, and in the process highlight key biological processes involved in DCM risk - including cellular adhesion, sarcomeric function, cell signalling, and ER stress.
2/5
21.11.2024 13:44 — 👍 1 🔁 0 💬 1 📌 0
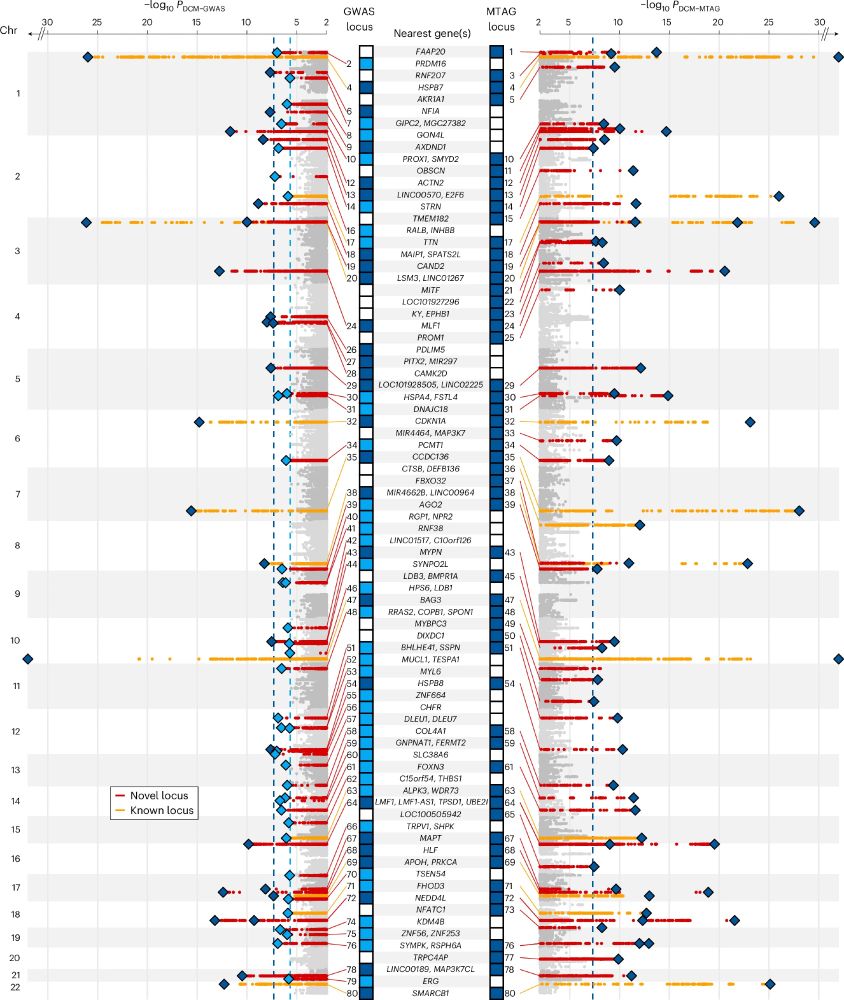
We analyse genetic data from >14K people with DCM and >1.2M controls, boosting power with multi-trait analysis incorporating CMR traits from 36K people. We find 80 risk loci (many novel) associated with DCM.
1/5
21.11.2024 13:41 — 👍 1 🔁 0 💬 1 📌 0
🧬 Our dilated #cardiomyopathy GWAS out today!
📍https://www.nature.com/articles/s41588-024-01952-y
All made possible with friends and collaborators from HERMES Consortium @alberthenry.bsky.social @tomlumbers.bsky.social @jamesware.bsky.social @mrc-lms.bsky.social @imperialnhli.bsky.social #BHF
🧵 👇
21.11.2024 13:35 — 👍 43 🔁 23 💬 5 📌 3
Mito & Heart Transplant Mom • Patient Advocate • Radiation Oncologist • Assistant Professor • Director of Wellbeing, Equity and Belonging • United Mitochondrial Disease Foundation (UMDF) Board of Trustees • Enduring Hearts Board Member
Sciencey boy working in coms
🌱🦠🏳️🌈
🇨🇦 in 🇬🇧. Assistant Prof. studying polygenic scores & multimorbidity at University of Cambridge; co-lead @pgscatalog.bsky.social. Otherwise, probably talking about good films, bad tv, or wine.
An open database of polygenic scores and the relevant metadata required for accurate application and evaluation
https://www.pgscatalog.org/
Medicine resident at Brigham and Women’s Hospital & genetics postdoc at the Broad Institute and Uni Helsinki. In Boston for the weather.
Bioinformatic scientist at work to disentangle the genetic basis of cardiovascular disease and mental illness 🧬🫀🧠
University of Brescia 🇮🇹 | Imperial College London 🇬🇧
Travel lover 🌏 | ⚽ player | 🏉 fan | many places to call 🏡
Bioinformatics | Data Science
Research, news, and commentary from Nature, the international science journal. For daily science news, get Nature Briefing: https://go.nature.com/get-Nature-Briefing
NHS Clinical Scientist. Inherited cardiac conditions. Genomics in healthcare. Oxford.
Co-Founder and Chief Scientist | Venture Partner | Professor | Mentor | Proud mom of 3 ✨
Researcher at Institute for Molecular Bioscience, The University of Queensland.
Genomics, Cardiovascular Disease
Lab group led by Prof Bernard Keavney focussing on cardiovascular genomics, big data & congenital heart disease in the @uom-bhfcre.bsky.social 🫀🧬
Posts by group members
🌐 https://www.keavneylab.com
📍 Manchester, UK
Writer, strategist, co-presenter the U.K. No1 podcast The Rest Is Politics … love Britain hate Brexit, loved Twitter loathe Musk n Trump
Broadcaster @lbc.co.uk
Author: https://www.waterstones.com/book/how-they-broke-britain/james-obrien/9780753560365
physician-scientist, author, editor
https://www.scripps.edu/faculty/topol/
Ground Truths https://erictopol.substack.com
SUPER AGERS https://www.simonandschuster.com/books/Super-Agers/Eric-Topol/9781668067666
Cardiology Registrar, Bristol Heart Institute
GW4-CAT PhD Fellow, Integrative Epidemiology Unit, Bristol
---
Cardiomyopathies, heart failure, genetics, obesity, arrhythmia and devices.