Of course, I would like to thank all of the members of FreyreLab for their cooperation and continuous support!
And thank you for reading! 😄
freyrelab.org/es/team
24.10.2025 19:10 — 👍 1 🔁 0 💬 0 📌 0
Finally, we conclude with a series of guidelines for improving the accomplishment of standards (such as the FAIR principles), as well as the availability of data and metadata.
24.10.2025 19:10 — 👍 0 🔁 0 💬 1 📌 0

On the other hand, we assess the availability of bacterial microarray data. Although this technology is being progressively displaced by RNAseq, it accounts for about half of the high-throughput gene expression records of bacteria, highlighting the importance of reevaluating all kinds of datasets.
24.10.2025 19:05 — 👍 1 🔁 0 💬 1 📌 0

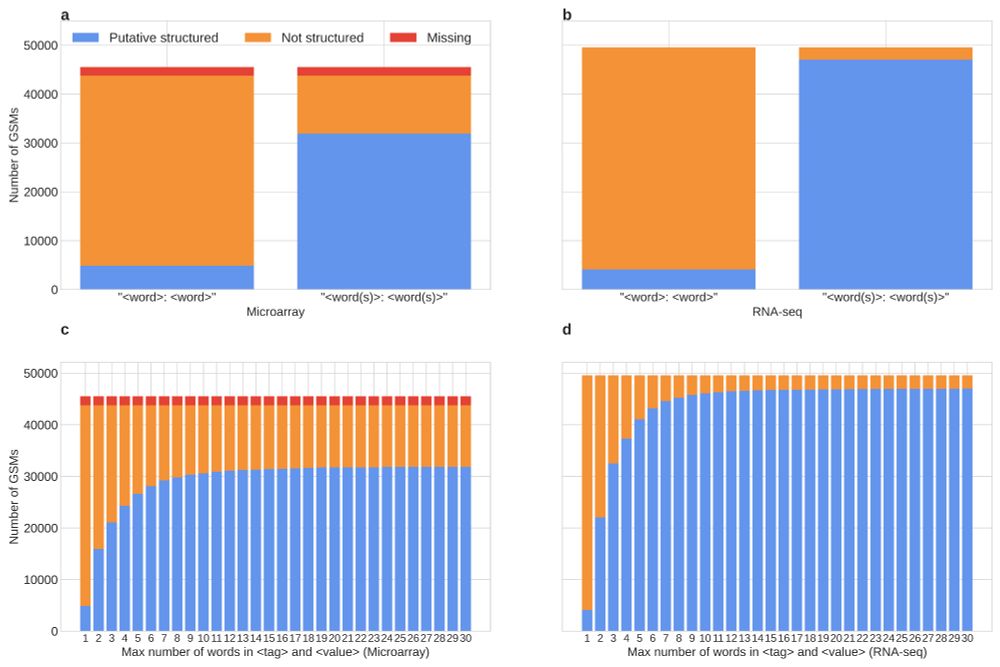
Regarding biological metadata, we analyse the adherence of the 'characteristics' fields to a <tag>:<value> structured format, as reported in the GEO documentation. Nevertheless, we found poor to mid adherence to this format, again limiting our possibilities for programmatic access to metadata.
24.10.2025 18:57 — 👍 0 🔁 0 💬 1 📌 0

In addition to those concerns, we report several inconsistencies in the GEO documentation that, we argue, strongly limit our potential to automatically extract metadata from GEO records.
24.10.2025 18:52 — 👍 0 🔁 0 💬 1 📌 0
However, this raises several concerns about data availability, reusability, and processing. In this perspective, we evaluate the severity of these issues in the GEO database, focusing on bacterial datasets.
24.10.2025 18:49 — 👍 0 🔁 0 💬 1 📌 0
The explosion in the amount of transcriptomics data (DNA microarrays and RNAseq) opens the possibility of harnessing publicly available datasets for big data applications to better understand gene regulation.
24.10.2025 18:47 — 👍 0 🔁 0 💬 1 📌 0
Evolutionary and conservation
genomics group leader at Globe institute, University of Copenhagen
🇲🇽in🇩🇰
https://globe.ku.dk/research/hologenomics/evolutionary-and-conservation-group/
Computational Biologist 🔬💻
Dog Person 🐶
he/him
GER/ENG
Group Leader @EMBLBarcelona | PD @TheCleversLab | PhD @MITBiology (Jacks lab) | B.A. @NewCollegeofFL | She/her | 🇵🇷🇺🇸🇪🇺| #Organoids, #Neuroendocrine cells
Evolutionary biologist | Head of the Evolutionary developmental biology lab @thecrick.bsky.social | Books, books, books!
We are the largest biomedical science hub in the south of Europe, by the beach in Barcelona.
Recerca biomèdica amb vistes |
@researchmar.bsky.social @crg.eu @melisupf.bsky.social @embl.org @isglobal.org @ibe-barcelona.bsky.social @fpmaragall.bsky.social
Research, news, and commentary from Nature, the international science journal. For daily science news, get Nature Briefing: https://go.nature.com/get-Nature-Briefing
Hola! We are the CRG, a research institute exploring the frontiers of biology in Barcelona. Som un centre CERCA. www.crg.eu
The European Molecular Biology Laboratory drives visionary basic research and technology development in the life sciences. www.embl.org
The Barcelona Collaboratorium for Modelling & Predictive Biology is a joint initiative by @crg.eu & @embl.org, exploring biology's frontiers with computational and theoretical modeling to make systems predictable & controllable.
The California Institute of Technology aims to expand human knowledge and benefit society through research integrated with education.
Official account for Harvard University. Devoted to excellence in teaching, learning, and research, and to developing leaders who make a difference globally.
#RStats @Bioconductor.bsky.social/🧠 genomics @lieberinstitute.bsky.social + Asst. Prof. @jhubiostat.bsky.social/@lcgunam @jtleek.bsky.social @aejaffe.bsky.social alumni/ @libdrstats.bsky.social @CDSBMexico.bsky.social
http://lcolladotor.github.io
Computational biologist at LIIGH-UNAM. Check my lab at https://sur.science
International Laboratory for Human Genome Research
Laboratorio Internacional de Investigación sobre el Genoma Humano
https://liigh.unam.mx/
@UNAM
🇲🇽 #RMB node: Community of Bioinformatics Software Developers #rstats @Bioconductor.bsky.social #bioinformatics
Admins @lcolladotor.bsky.social @areyesq.bsky.social @nnb_unam
Group Leader at LIIGH-UNAM | yeast genomics | hybridization | microbial communities
PhD student @crg.eu Interested in #bioinformatics #genomics #generegulation #transposons #evolution #popgen
Genómico 🧬💻🧬
Biología de sistemas evolutivos | LIIGH | UNAM
Doctorante de Ciencias Biomédicas




