Further, because COAST-SS operates on summary statistics instead of individual-level genotypes, it can complete a genome-wide scan 100x faster than the original COAST.
18.10.2025 11:57 — 👍 2 🔁 0 💬 1 📌 0
Of the 108 candidate allelic series identified in our analysis, 80 had prior rare variant support, and 23 of the remaining 28 associations had common variant support. The remaining 5 associations, all with HDL, are reported here for the first time.
18.10.2025 11:57 — 👍 1 🔁 0 💬 1 📌 0

In a combined analysis of UKB and MVP with a sample size of up to 840K, we identify 108 Bonferroni-significant genes associated with lipid traits, including 41 for HDL (below), more than doubling the number of genes identified in our previous analyses.
18.10.2025 11:57 — 👍 1 🔁 0 💬 1 📌 0

Though we recommend using LD calculated in a relevant sample, due to the low LD among rare variants, the test remains valid even when variants are assumed to be in linkage equilibrium.
18.10.2025 11:56 — 👍 1 🔁 0 💬 1 📌 0

In simulations and real data, we show that COAST-SS is robust to how linkage disequilibrium (LD) is specified.
18.10.2025 11:55 — 👍 1 🔁 0 💬 1 📌 0
Here we present COAST-SS, which enables detection of allelic series genes based entirely on per-variant GWAS summary statistics.
18.10.2025 11:55 — 👍 1 🔁 0 💬 1 📌 0

A scalable framework for identifying allelic series from summary statistics
Genes where a dose-response relationship exists between functionality and phenotypic impact are appealing therapeutic targets, as the effects of pharm…
We’re excited to share a new article from @insitro.bsky.social (first author @zmccaw.bsky.social) in @ajhgnews.bsky.social, introducing a summary-statistic-based implementation of the COding-variant Allelic Series Test, COAST-SS: www.sciencedirect.com/science/arti...
18.10.2025 11:53 — 👍 6 🔁 4 💬 1 📌 0
Today insitro announces an exciting new collaboration with insighteyehub.bsky.social at Moorfields Eye Hospital, London, through the development of a novel #AI foundation model to identify and to support #drugdiscovery. Learn more: www.businesswire.com/news/home/20...
18.03.2025 10:06 — 👍 3 🔁 2 💬 0 📌 2
@tkaraletsos.bsky.social, Daphne Koller, Hugues Aschard, @mendelrandom.bsky.social, @dgmacarthur.bsky.social, @codushlaine.bsky.social, and additional members of the team at @insitro.bsky.social !
11.02.2025 12:46 — 👍 3 🔁 0 💬 1 📌 0
Congrats to the team and our collaborators! @zmccaw.bsky.social, Rounak Dey, Hari Somineni, David Amar, @smukherjee89.bsky.social, Kaitlin Sandor, ...
11.02.2025 12:45 — 👍 4 🔁 0 💬 1 📌 0

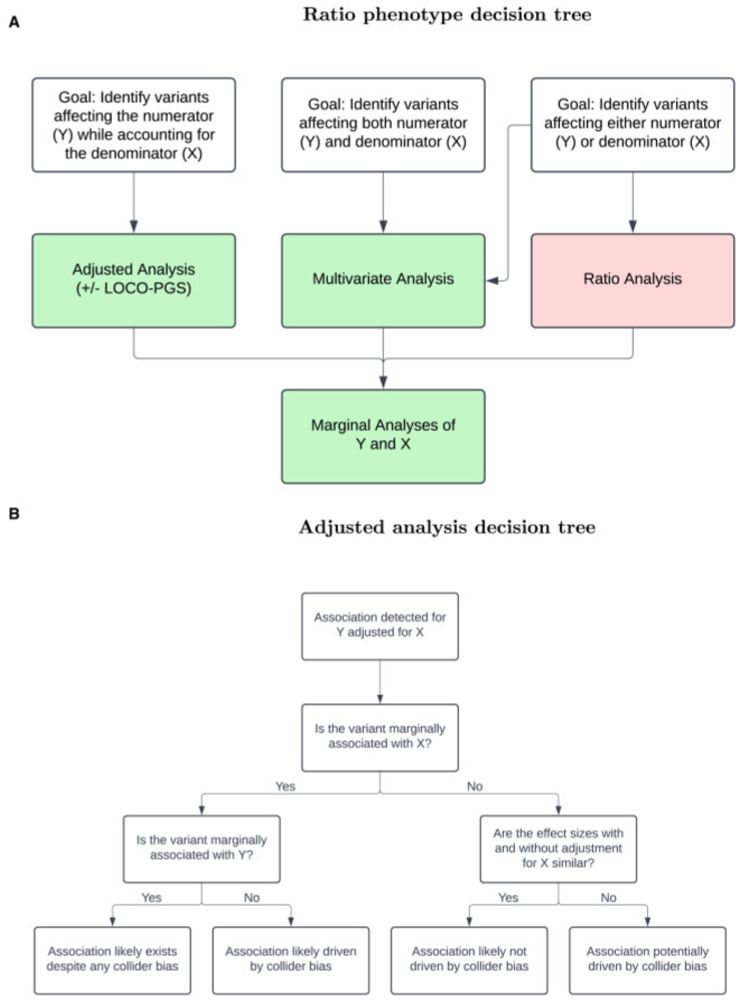
In summary, associations with ratio traits are not specific to the ratio per se. Practitioners considering GWAS on ratios should examine the motivation for forming the ratio. We provide guidance on how to approach GWAS of related pairs of traits in light of analysis objectives.
11.02.2025 12:43 — 👍 2 🔁 0 💬 1 📌 0

In the case where the denominator is heritable and collider bias is a concern, we propose additionally adjusting for a leave-one-chromosome-out polygenic score (LOCO-PGS) of the denominator, which we show corrects for collider bias due to genetic background.
11.02.2025 12:43 — 👍 1 🔁 0 💬 1 📌 0
Instead of a ratio model, we propose a model that adjusts for the denominator as a covariate, e.g. to detect the genetic effect on weight while holding height constant. Loci from weight adjusted for height GWAS were enriched for adiposity-related signals more so than loci from the BMI GWAS.
11.02.2025 12:42 — 👍 1 🔁 0 💬 1 📌 0

Examining ratio traits commonly used in clinical practice and genetic association testing, such as waist-to-hip ratio and albumin-to-creatinine ratio, we find that loci detected by the ratio model are enriched for denominator signals.
11.02.2025 12:41 — 👍 2 🔁 0 💬 1 📌 0

Top “hits” from ratio trait GWASs can be driven entirely by the denominator. This means, for example, that attempts to find genes associated with BMI may return genes associated entirely with height (only), and not weight.
11.02.2025 12:40 — 👍 2 🔁 0 💬 1 📌 0
With high-dimensional omics and imaging data, there is a temptation to test all pairs of ratios to increase power. We caution against this approach; GWAS on ratio traits may increase the number of significant associations, but it does so by conflating signals for the numerator and denominator.
11.02.2025 12:39 — 👍 2 🔁 0 💬 1 📌 0
Medicinal chemist / chemical biologist, author of “In the Pipeline” at http://science.org/blogs/pipeline. derekb.lowe@gmail.com and on Signal at Dblowe.18
All opinions are mine; I don’t speak for my employer in any way.
If Adolf Hitler flew in today, they’d send a limousine anyway. Parody. Merch: https://tinyurl.com/msjjmwpt
Dad, husband, President, citizen. barackobama.com
Bluesky newbie. Twitter exile. Previously Professor of Diabetes in Oxford, now heading human genetics at Genentech. Living the California dream. Views my own. #YNWA
Temerty Professor of Modern European History, Munk School, University of Toronto; Permanent Fellow, IWM Vienna; Emeritus Levin Professor, Yale. Author of "On Freedom," "On Tyranny," "Our Malady," "Road to Unfreedom," "Black Earth," and "Bloodlands"
DC. political philosopher at Georgetown. rhythm guitarist + vox for @femiandfoundation.bsky.social. Spurs fan #COYS
Psychology & behavior genetics. Author of THE GENETIC LOTTERY (2021) and ORIGINAL SIN (coming 3.3.2026). Speaking as an individual
Statistical Geneticist, Group leader at University of Lausanne/Unisante, father, climber, runner
Researcher at University of Lausanne | interested in genetic epidemiology, mental health & behaviour
PubPeer is a platform for post-publication peer review, offering an additional layer of evaluation beyond journal names. Empowering the scientific community to assess science and scientists transparently. pubpeer.com
All views and opinions expressed are my own.
Postdoctoral Fellow with Dr. Greg Gibson, Center for Integrative Genomics, Georgia Tech, Atlanta, GA
PhD Bioinformatics (Statistical Genetics) | MS Bioinformatics @GeorgiaTech
https://sininagpal.wixsite.com/snagpal
yes, really the Northeast Ohio Regional Sewer District in cleveland, ohio. Customer Relations: 216.881.8247
https://www.neorsd.org/social
Official account for Rep. Moulton's press office. Member of House Armed Services Committee, Committee on Transportation and Infrastructure, and Select Committee on the CCP
Know Your Rights https://moulton.house.gov/immigration-rights
U.S. Senator, Massachusetts. She/her/hers. Official Senate account.
https://substack.com/@senatorwarren
U.S. Senator fighting for the families and future of Massachusetts. Malden Native, Red Sox Fan, Net Neutrality Defender, co-author of the Green New Deal ( he/him )
Senator for Massachusetts. Ranker on Senate Small Business Committee & HELP Primary Health & Retirement Security Subcommittee. Fighting for a Green New Deal.
Local Indivisible chapter. Figuring out how to do the work, one messy moment at a time.
EMBO Postdoctoral fellow at the Garvan Institute of Medical Research, Sydney, Australia.
Previously EMBL-EBI, Wellcome Sanger Institute and University of Cambridge in Cambridge, UK.
All things single-cell, genetics & genomics.