
Thread here @anshulkundaje.bsky.social :
x.com/Akhiad6/stat...
We'll try to update on comparisons and ensembles with the one and only ChromBPNet
@akhiad.bsky.social

Thread here @anshulkundaje.bsky.social :
x.com/Akhiad6/stat...
We'll try to update on comparisons and ensembles with the one and only ChromBPNet

A biophysically principled sequence model IceQream characterises complex Transcription Factor-DNA interactions from chromosome accessibility analysis.
@Akhiad.bsky.social
#BiotechNatureComms
www.nature.com/articles/s41...
Huge props to CREsted (@niklaskemp.bsky.social & @steinaerts.bsky.social ) & Borzoi (Johann Linder & David R. Kelley) creators for enabling the DL benchmarking.
Learn more on IQ:
Paper:
www.nature.com/articles/s41...
GitHub:
github.com/tanaylab/ice...
Analysis code:
github.com/tanaylab/IQ-...
15/
Working with @aviezerl.bsky.social has been a blast, the wonderful Roni Stok & Saifeng Cheng (with Yonatan Stelzer’s guidance) conducted the experiments and data collection, with all efforts orchestrated and led by Amos Tanay.
14/
IQ lays a foundation for modeling other epigenomic features. We're using it to study Polycomb domains, DNA methylation and insulation. We optimistically believe all epigenomic aspects can be combined to model differentiation programs from sequence alone.
13/

We are also curious to learn new biology when black box models outperform IQ. In CRE sequences alone, LLMs have not yet revealed grammars that transcend IQ’s simple TF-DNA interactions. Longer-range chromosomal interactions among CREs, we believe, may be a different story.
12/

We are excited about the possibility of using IQ to develop better DL for epigenomes. We do gain from using ensembles of IQ and DL models – so there is hope! LLMs consider larger contexts than IQ, while IQ is super economical in parameters and may generalize better because of that.
11/

Insight 3: Local interactions between TFs. IQ predicts TF interactions that predict CRE accessibility across differentiation trajectories. For example: Mesp-Eomes motif co-occurences may be important for germ-layer specification.
10/

Insight 2: TFs care about sub-optimal binding sites. IQ integrates strong and weak TF-DNA interactions. This reveals that TFs 'read' sequences in different ways – some care only for the best targets and others integrate many weak ones.
9/

Here are 3 biological insights derived from IQ (before we reflect on what’s in it for the non-biologist crowd).
Insight 1: Sequence defines regulatory intensity over a quantitative spectrum – not a binary yes/no classifier. IQ can predict this spectrum!
8/

For example, when modelling regulation of Epiblast to Mesoderm differentiation in mouse embryos, we can explain changes in accessibility with only 13 TF motif models!
7/

IQ inference starts from detailed model and progressively simplifies it to a small set of physical models. Models with smaller number of components are easier to interpret. They also generalize better.
6/

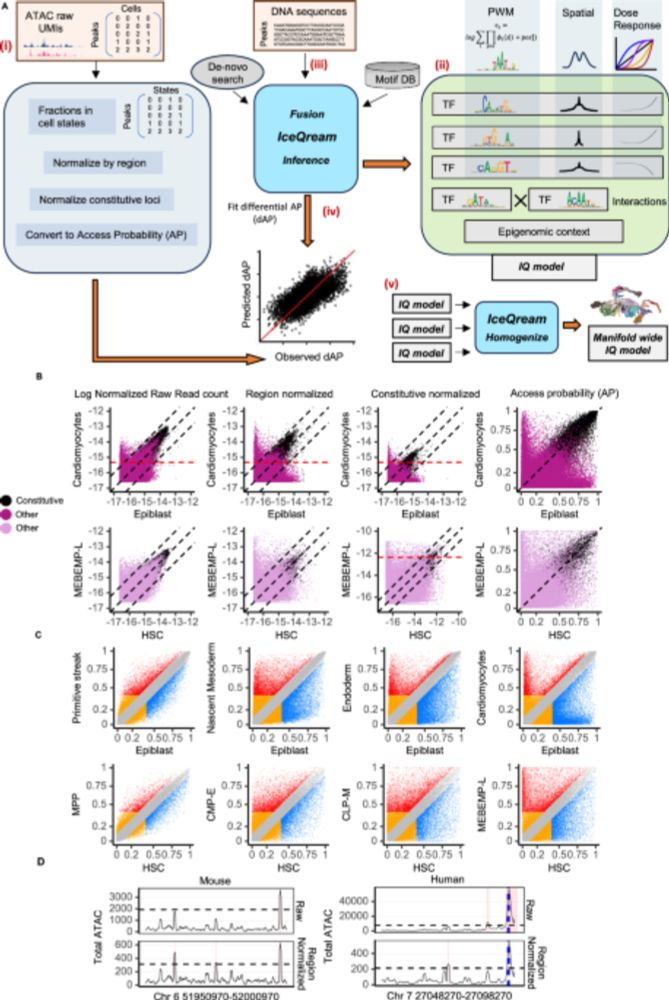
IQ regresses AP from sequence using biophysically inspired TF binding models including:
*Spatial integration across a range of TF-DNA affinities
*Latent TF concentrations with non-linear dose-response
*Synergistic/antagonistic pairwise TF interactions
5/

IQ uses a normalization trick based on coverage of constitutive ATAC peaks to derive APs robustly. Because AP is defined on an absolute scale – comparing AP among conditions is immediate and robust.
4/

But IQ is more than a predictive tool. A first important difference between IQ and other approaches is the transformation of ATAC-seq coverage to access probabilities (AP) - the instantaneous chances to find a CRE in an open state among cells from a given type.
3/

As a mere predictive tool, IQ performs on par with state-of-the-art DL models such as Borzoi and DeepTopic. Evaluation on human blood and mouse embryo datasets shows that current best performance is derived using an ensemble of IQ and Borzoi.
2/

Do we need LLMs to predict epigenomes from DNA—or is biophysics enough? 🧬
IceQream (IQ) is a biophysics-based framework that predicts epigenomes with SOTA-level accuracy—and is fully explainable.
@NatureComms: www.nature.com/articles/s41...
Thread 🧵👇
1/