
Relieved to finally post my whole developing brain evolutionary "theory of everything" preprint!
www.biorxiv.org/content/10.1...
@niklaskemp.bsky.social
PhD Student at the Stein Aerts Lab of Computational Biology. Studying brain genomics

Relieved to finally post my whole developing brain evolutionary "theory of everything" preprint!
www.biorxiv.org/content/10.1...
We have two open positions for a ML and a LLM engineer to launch a machine learning expertise unit in our center @vibai.bsky.social, see vib.ai/en/opportuni...
23.09.2025 11:21 — 👍 5 🔁 8 💬 0 📌 0We will have our next community meeting on Tuesday, 2025-09-16 at 18:00 CEST! Niklas Kempynck will be presenting on CREsted, a package for training enhancer models on scATAC-seq data.
(Zoom registration link and more information in thread!)
🧵

I wrote a quick application note on Tomtom-lite, a Python implementation of the Tomtom algorithm for comparing PWMs against each other. This implementation can be 10-1000x faster and, as a Python function, can be integrated into your workflows easier.
www.biorxiv.org/content/10.1...
One thousand candidate enhancers tested in vivo in the mouse brain! A massive resource and oh so useful as validation set for genome-wide enhancer prediction methods. Super fun to be involved in one of the papers: ‘the prediction challenge paper’ by Nelson&Niklas et al www.cell.com/cell-genomic...
21.05.2025 16:50 — 👍 43 🔁 13 💬 0 📌 0Make sure to also check out the other studies part of the larger effort on identifying and validating enhancer tools.
21.05.2025 16:51 — 👍 0 🔁 0 💬 0 📌 0This study was done together with Nelson Johansen and supervised by Trygve Bakken at the @alleninstitute.org. Thanks to all co-authors for the great inter-lab collaboration! Also a personal shoutout to the members in @steinaerts.bsky.social lab for a nice team effort and to Stein for guidance.
21.05.2025 16:48 — 👍 2 🔁 0 💬 1 📌 0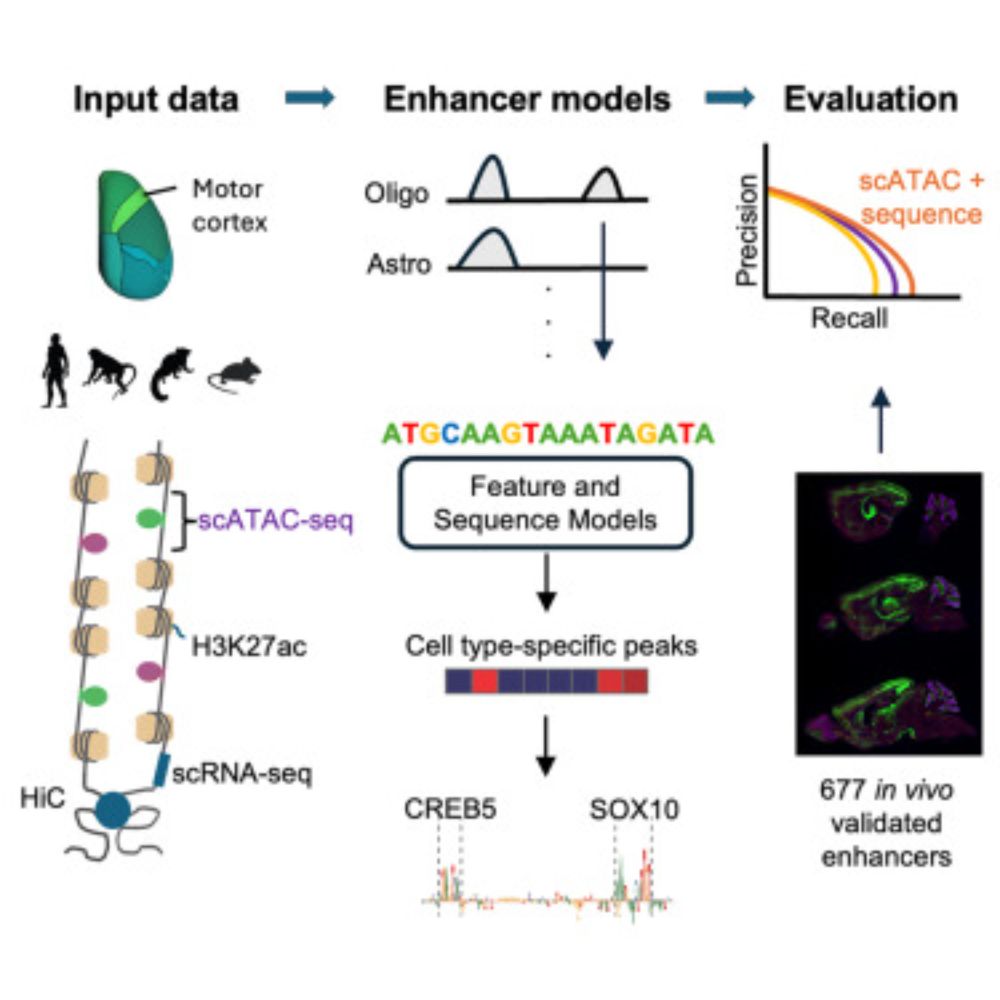
Check out our work on evaluating methods for predicting in vivo cell enhancer activity in the mouse cortex! Combined, scATAC peak specificity and sequence-based CREsted predictions gave the best predictive performance, aiming to advance genetic tool design for cell targeting in the brain.
21.05.2025 16:45 — 👍 20 🔁 10 💬 1 📌 0
Calling someone bird-brained is, in fact, a way of calling someone highly intelligent. @yaseminsaplakoglu.bsky.social reports: www.quantamagazine.org/intelligence...
07.04.2025 14:28 — 👍 92 🔁 26 💬 2 📌 5
Very proud of two new preprints from the lab:
1) CREsted: to train sequence-to-function deep learning models on scATAC-seq atlases, and use them to decipher enhancer logic and design synthetic enhancers. This has been a wonderful lab-wide collaborative effort. www.biorxiv.org/content/10.1...
Also check out Hannah’s thread on our latest preprint on HyDrop v2, an open-source platform for scATAC-sequencing, and a great, cost-efficient way of generating data for S2F models. 🙌
04.04.2025 10:37 — 👍 7 🔁 1 💬 0 📌 0
CREsted is available at github.com/aertslab/CRE.... Analysis notebooks can be found at github.com/aertslab/CRE.... All models developed for this preprint and in previous work are available in CREsted through crested.get_model(). We look forward to your feedback!
03.04.2025 14:36 — 👍 2 🔁 1 💬 0 📌 0This was a big collaborative effort, together with @seppedewinter.bsky.social , and with great contributions from @casblaauw.bsky.social , Vasilis and many others. A special shoutout to @lukasmahieu.bsky.social who professionalized the package, and to @steinaerts.bsky.social for supervising.
03.04.2025 14:35 — 👍 1 🔁 0 💬 1 📌 0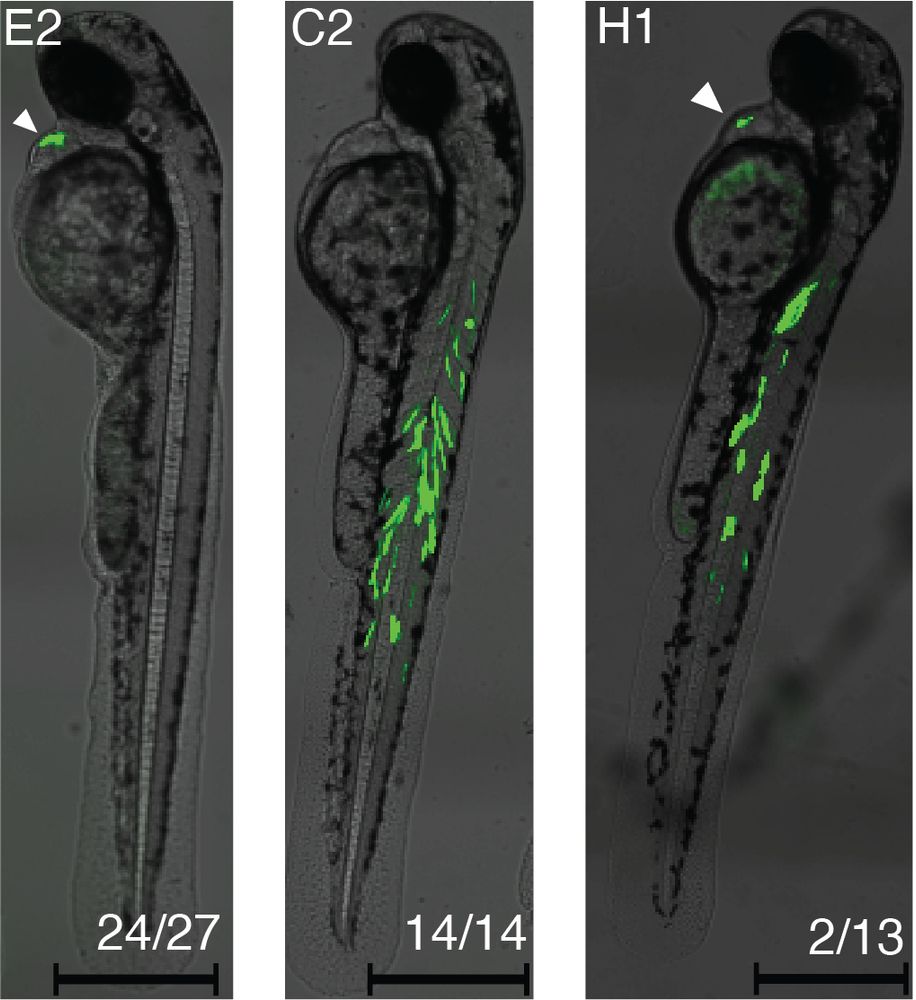
Finally, we train a model on a full-development zebrafish scATAC-seq atlas, and use it to design and in vivo validate cell type- and timepoint-specific enhancers with a high success rate. We also attempt to modulate reporter strength over two cell types.
03.04.2025 14:34 — 👍 3 🔁 0 💬 1 📌 0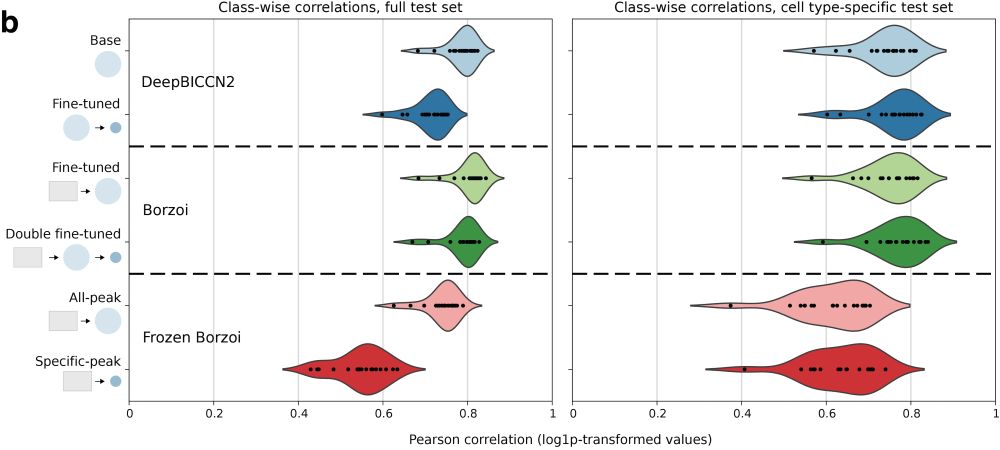
In a new functionality to CREsted, we explore Borzoi fine-tuning to mouse motor cortex scATAC-seq data. We show that fine-tuned models and smaller models from scratch have a near-identical performance.
03.04.2025 14:34 — 👍 1 🔁 0 💬 1 📌 0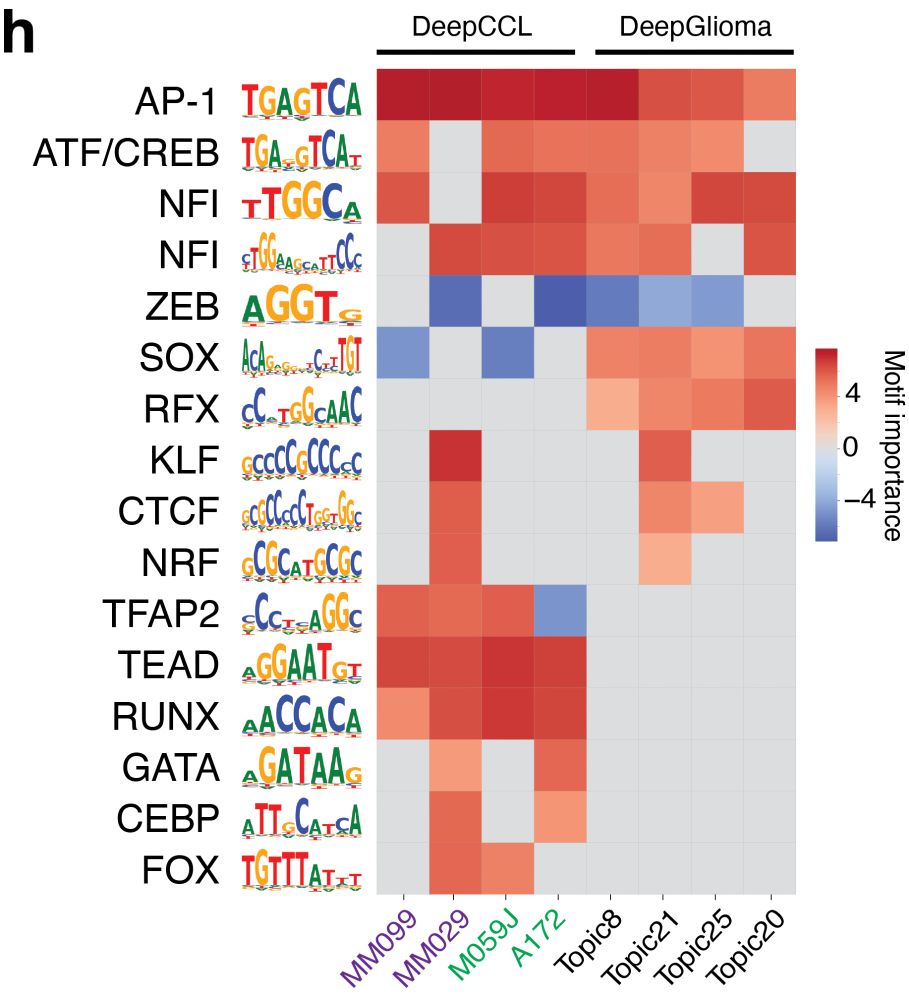
We also study enhancer code inside human cancer cell lines and glioma biopsies and find that enhancer codes between Mesenchymal-like glioblastoma and melanoma states are more similar compared to glioblastoma biopsy data.
03.04.2025 14:33 — 👍 1 🔁 0 💬 1 📌 0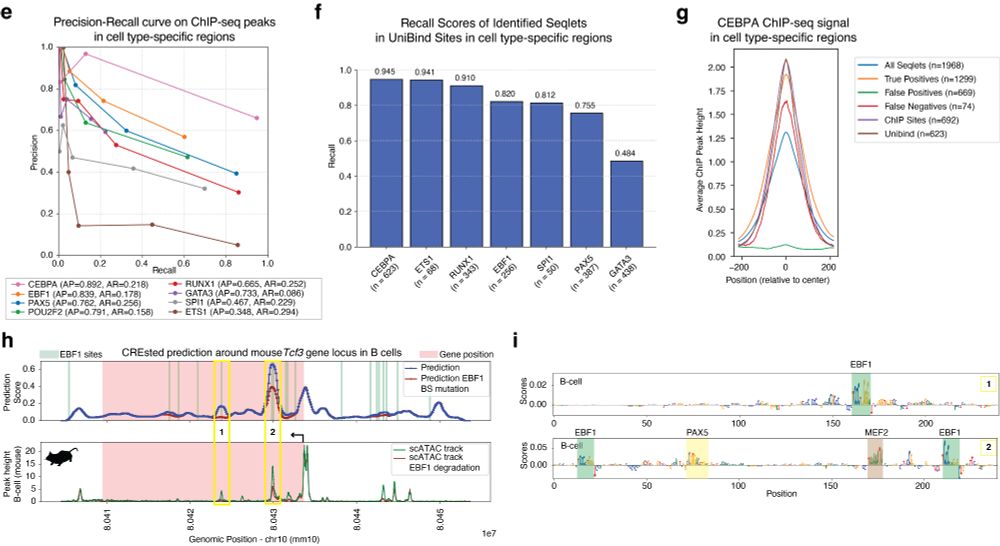
Next, we validated CREsted-identified motif instances from a human PBMC model with ChIP-seq data. We further show that gene locus predictions can be used to simulate the effect of TF degradation on chromatin accessibility.
03.04.2025 14:32 — 👍 1 🔁 0 💬 1 📌 0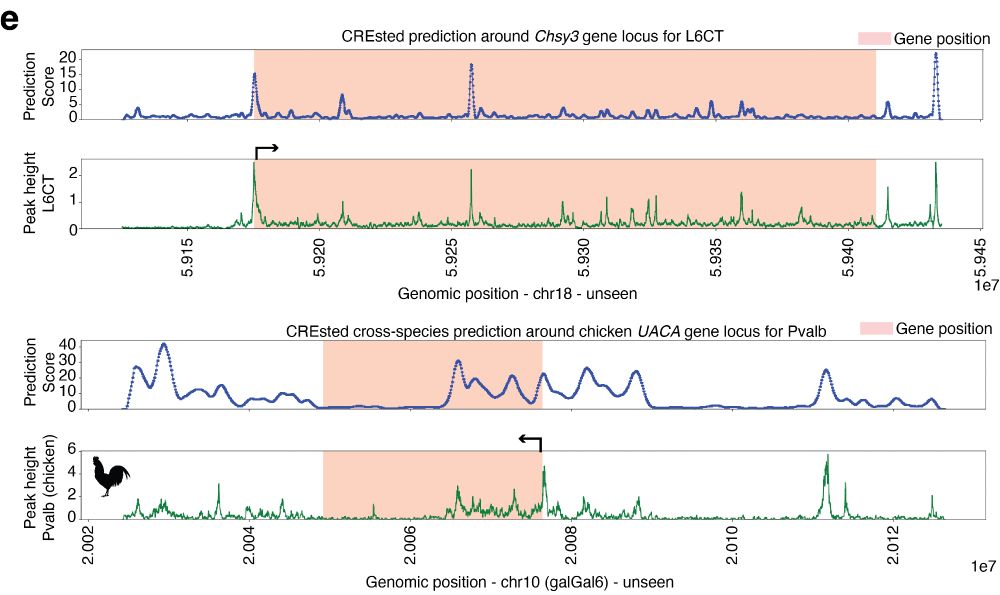
We use the mouse cortex model to highlight CREsted’s gene locus prediction capabilities, both in unseen chromosomes and across species. This presents a powerful tool for potentially annotating genomes across species at high resolution.
03.04.2025 14:32 — 👍 1 🔁 0 💬 1 📌 0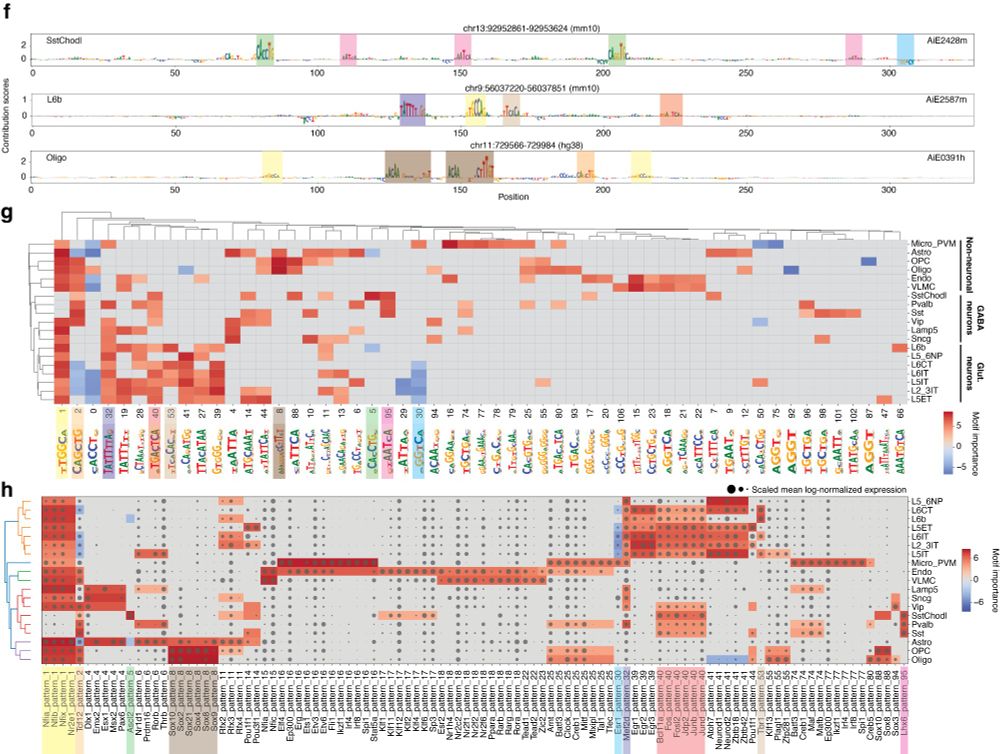
We first demonstrate CREsted’s functionality by providing a complete data-driven analysis of mouse motor cortex enhancer codes across cell types. Through matched scRNA-seq data, we link motifs to likely TF candidates.
03.04.2025 14:31 — 👍 2 🔁 0 💬 1 📌 0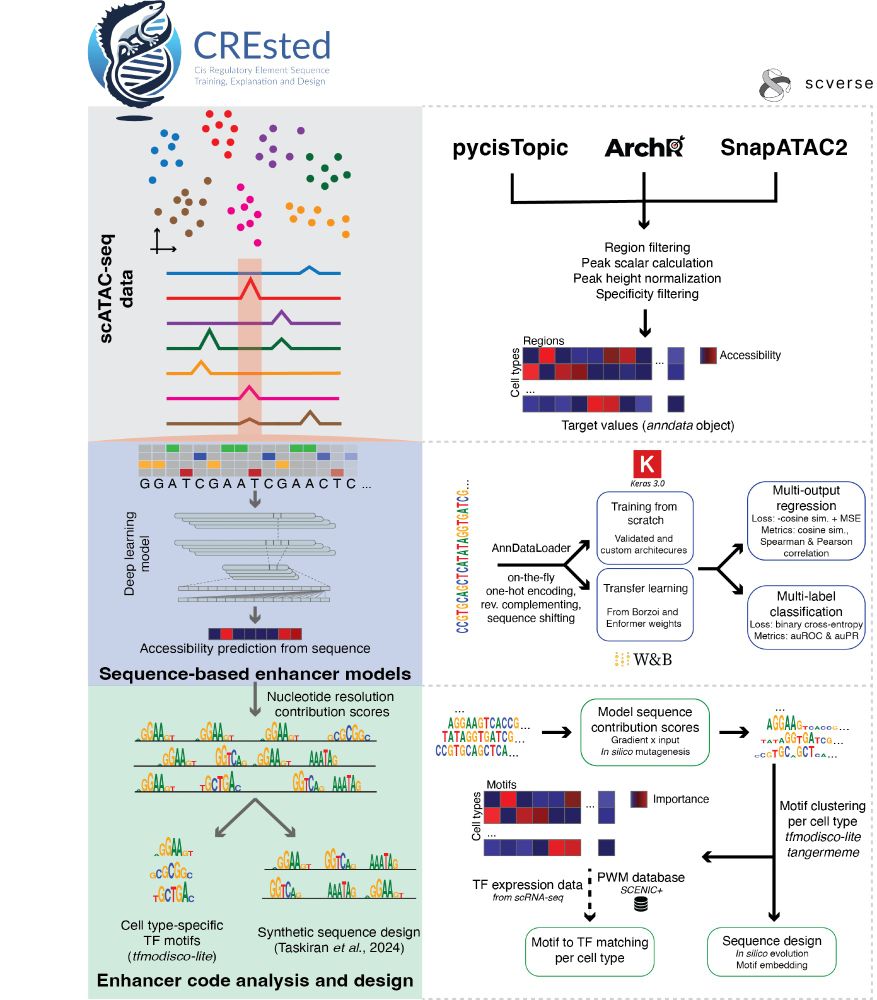
CREsted starts from the outputs of established scATAC preprocessing pipelines, and trains sequence-to-function models on chromatin accessibility per cell type. It provides complete motif analysis tools to infer cell type-specific enhancer codes and holds a comprehensive
enhancer design toolbox.

We released our preprint on the CREsted package. CREsted allows for complete modeling of cell type-specific enhancer codes from scATAC-seq data. We demonstrate CREsted’s robust functionality in various species and tissues, and in vivo validate our findings: www.biorxiv.org/content/10.1...
03.04.2025 14:30 — 👍 75 🔁 38 💬 1 📌 5Very excited to share our new preprint together with @daniedaaboul.bsky.social, where we studied the gene regulatory code that hippocampal granule cells (GCs) use during synapse formation (1/n)
31.03.2025 05:57 — 👍 15 🔁 8 💬 2 📌 0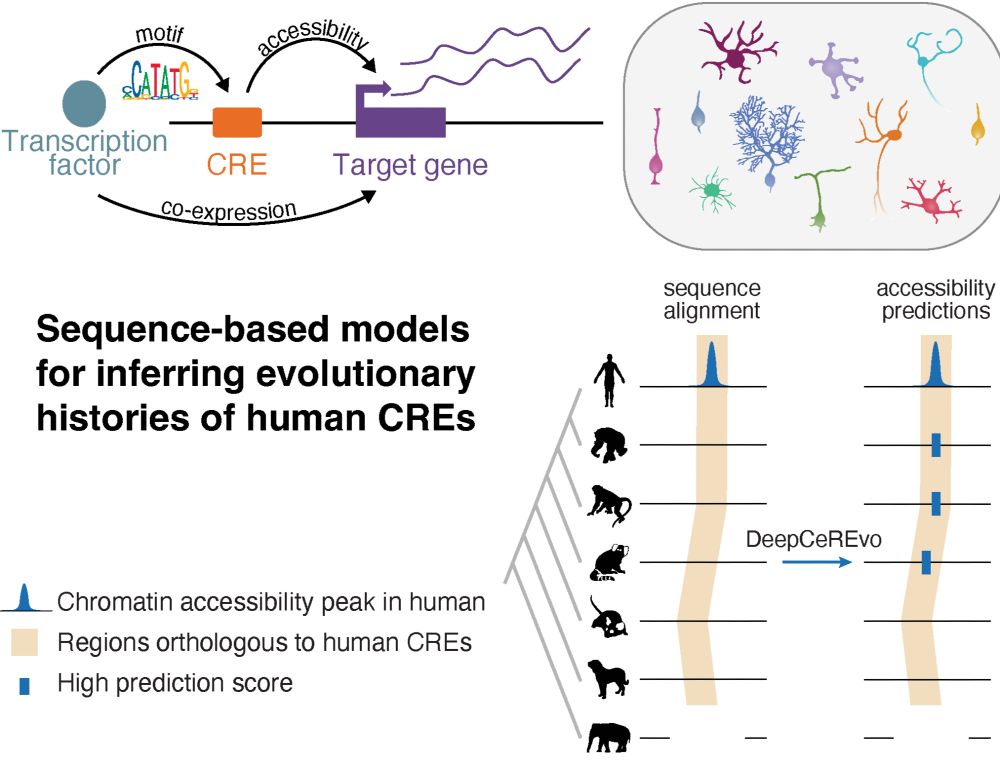
How does gene regulation shape brain evolution? Our new preprint dives into this question in the context of mammalian cerebellum development! rb.gy/dbcxjz
Led by @ioansarr.bsky.social, @marisepp.bsky.social and @tyamadat.bsky.social, in collaboration with @steinaerts.bsky.social

The latest Discover ASAP episode dives into "Cell Type Directed Design of Synthetic Enhancers," a study published in Nature by CRN Team Voet. They discuss how machine learning enables precise enhancer design for targeted gene expression 🧬
Watch: www.youtube.com/watch?v=Qcms...
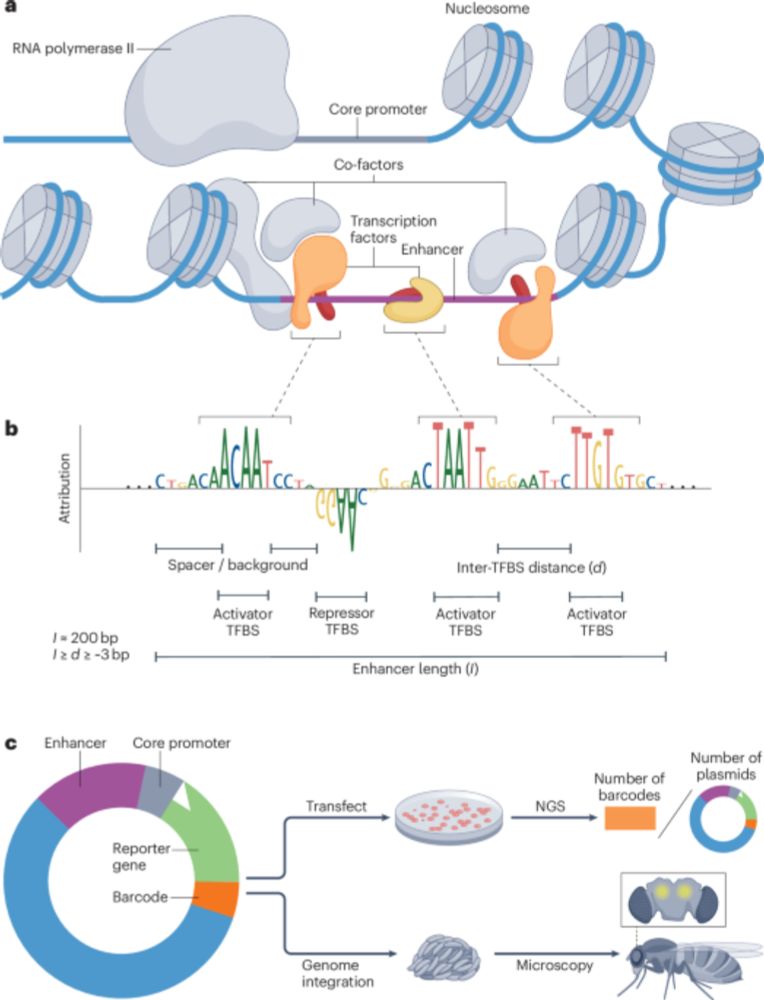
We wrote a review article on modelling and design of transcriptional enhancers using sequence-to-function models.
From conventional machine learning methods to CNNs and using models as oracles/generative AI for synthetic enhancer design!
@natrevbioeng.bsky.social
www.nature.com/articles/s44...
This has been a fantastic adventure - to capture the genomic regulatory code underlying brain cell types (using deep learning models trained on chromatin accessibility), and then use these models to compare cell types between the bird and mammalian brain
14.02.2025 12:06 — 👍 41 🔁 12 💬 4 📌 1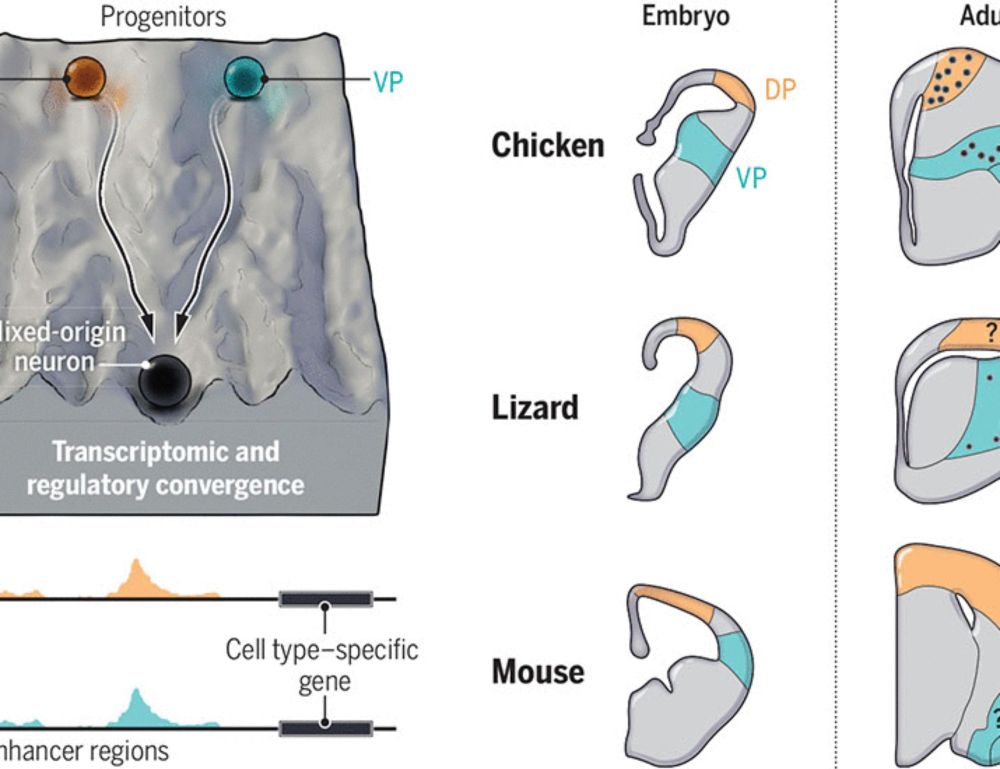
Also, check out the two related articles from the @kaessmannlab.bsky.social and García-Moreno groups, and the expert perspective by @giacomogattoni.bsky.social and Maria Antonietta Tosches www.science.org/doi/10.1126/...!
14.02.2025 10:13 — 👍 5 🔁 0 💬 0 📌 0Just very happy to have our paper out today! A big thanks to all our co-authors, and to Nikolai and @steinaerts.bsky.social for the teamwork over the past years. If you are interested in using our models for cross-species enhancer studies, check out crested.readthedocs.io/en/stable/mo... 🙂
14.02.2025 10:07 — 👍 53 🔁 25 💬 3 📌 3