Ding Lab
website description
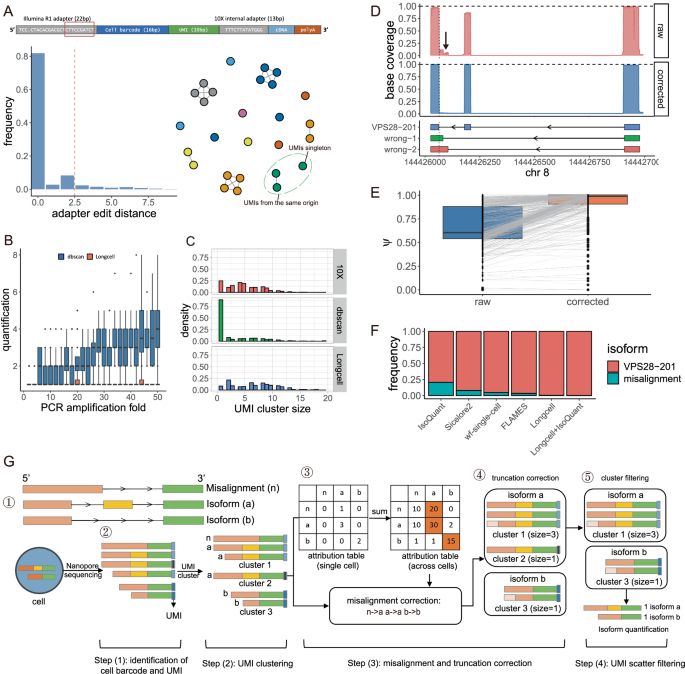
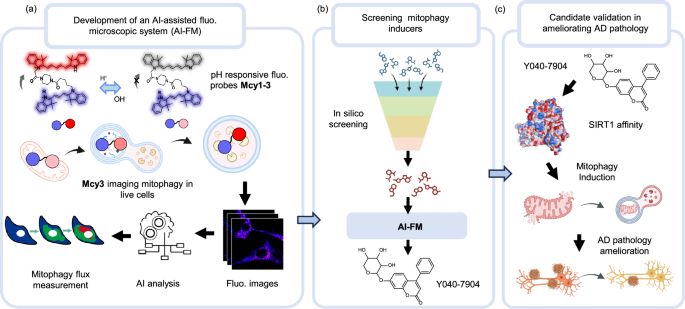
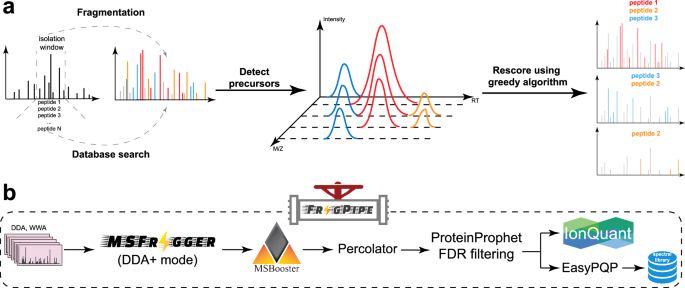
DOLPHIN: a deep learning method that enables exon- and junction-level analysis to improve cell representation and detect alternative #splicing. Ding Lab junding.lab.mcgill.ca #BiotechNatureComms
doi.org/10.1038/s414...
07.07.2025 15:09 — 👍 1 🔁 0 💬 0 📌 0

Green Laboratory
Visit the post for more.
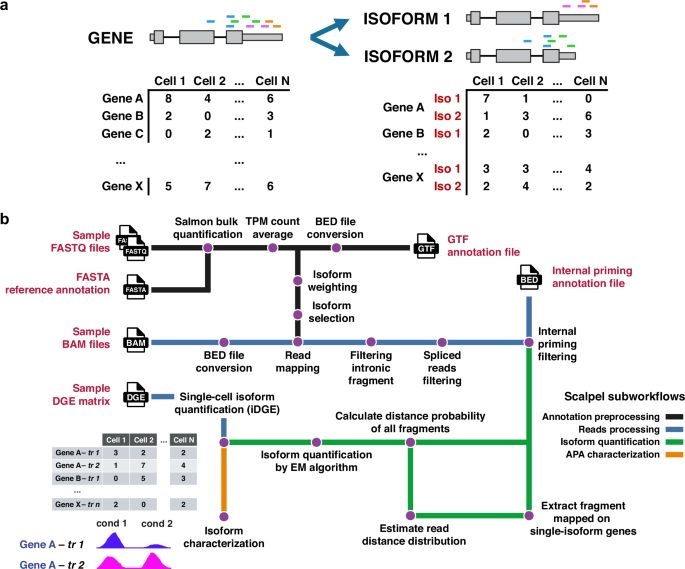
A data-efficient, generative AI-based toolkit for designing a diverse set of RNA molecules. #BiotechNatureComms
www.nature.com/articles/s41...
alexgreenlab.org
08.05.2025 13:29 — 👍 0 🔁 0 💬 0 📌 0

DNA-Based Computing and Data Storage
This cross-journal Collection shines a spotlight on research exploring DNA as a scalable and sustainable medium for computing and data storage.
🧬💻Call for papers: DNA-based Computing and Data Storage
@naturecomms.bsky.social @commsbio.bsky.social @commseng.bsky.social
@natureportfolio.nature.com
The deadline is mid September 2025. Feel free to drop me a message if you want to discuss submissions.
www.nature.com/collections/...
06.03.2025 07:05 — 👍 7 🔁 5 💬 0 📌 0
Leading research publisher empowering researchers, educators, and health professionals with our trusted brands for 180+ years. #BePartOfProgress
https://group.springernature.com/gp/group
Site notice http://bit.ly/2oAyLi1
Biophysicist & AI enthusiast. Professor at UVic-UCC. Group leader at QuBI lab.
Communications AI & Computing is an open access journal in the @natureportfolio.nature.com committed to the timely publication of impactful research in all areas of artificial intelligence and computational science.
www.nature.com/commsaicomp/
Systems & Synthetic Biology | Machine Learning | Control Theory | Applied Mathematics
https://homepages.inf.ed.ac.uk/doyarzun/
Oscillating between ticks and tocks. Circadian computational biologist now moving to the other side of Science, starting a new editorial career in Nature Communications
Aiming to make the Solar Punk Aesthetic a reality. Science, World Building and Story telling. Researcher/Bioinformatician/Data Scientist.
"Always strive to contribute to others."
Contact:
https://patrickcnmartin.github.io/
Postdoctoral Scientist at the University of Oxford. Interested in platelet and cancer biology 🩸
http://www.linkedin.com/in/lauren-c-murphy
Associate Professor, University of Pittsburgh. Department of Computational and Systems Biology. Lab of Cellular Dynamics and Molecular Circuits https://csbweb.csb.pitt.edu/Faculty/lee/
Dedicated to drug discovery, enthralled by science. 5 kids. She. SAB Recursion. Founder SyzOnc. Lab at Broad Institute. Opinions my own.
Editor of Nature Photonics. Based in London.
Group leader - computational biomedical discovery. Heidelberg University & Heidelberg University Hospital. https://www.tanevskilab.org
computational scientific discovery, biomedicine, spatial omics
PI of the machine learning for Integrative genomics lab @pasteur.fr and @CNRS.bsky.social
#NewPI #DiversityEqualityInclusion
Single-cell, multi-omics, machine learning
Editorial Director, Nature portfolio. Ex-Chief editor at Nature Microbiology & Nature Reviews Microbiology. Ex-Senior editor at Nature Biotechnology & PLOS Medicine. Microbiology& technology are my happy place. Here for the science, views my own.
Chief Editor of Nature Physics. Former runner 🏃. Wannabe crossfitter 🏋️. Mediocre photographer 📷. Opinions my own. Debate welcomed.
Senior Editor at Nature Communications handling Mol Bio & Genetics • 🇮🇪 in 🇬🇧 • Recovering academic with a soft spot for Drosophila • Also enjoys running 🏃♀️ and coffee ☕️ • All opinions my own
Group of Adrien Hallou @kiroxford.bsky.social @ox.ac.uk
Biophysics & spatial biology of cell fate decisions & tissue dynamics
Alumnus @cam.ac.uk & @normalesup.bsky.social
Franco-British Young Leader 2024 🇨🇵🇬🇧
Neuroscientist, Nature Neuroscience (@natneuro.nature.com) Chief Editor, Upstate NY mama of 2. Opinions are my own.
Rectangular gifts are easiest to wrap. https://www.amazon.co.uk/stores/Henry-Gee/author/B001IO9QAC?ref=ap_rdr&isDramIntegrated=true&shoppingPortalEnabled=true
Senior Publisher at the Nature Partner Journals series. Microbiology and bicycle as transport fan, climate change worrier. He/Him.
My journals: npj Biodiversity, npj Drug Discovery, npj Metabolic Health and Disease, npj Veterinary Sciences & npj Viruses