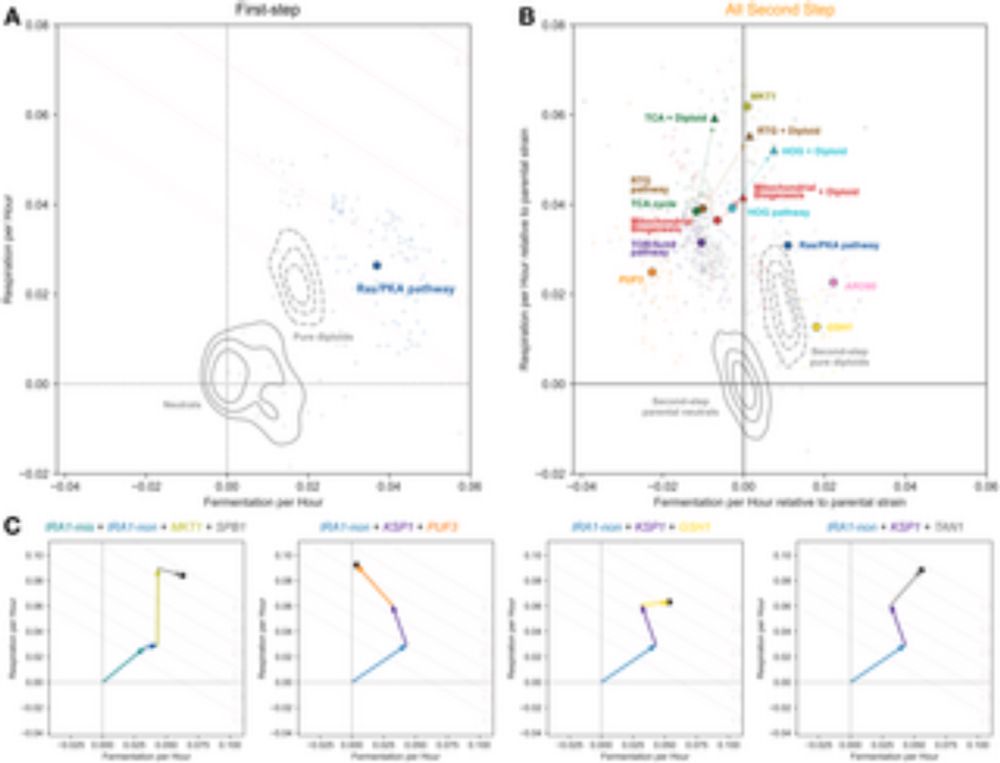
Super excited that the bulk of my PhD work is now preprinted! Here we used whole-community competition, or coalescence, experiments to quantify selection acting on genetically diverged strains within larger communities. (1/n)
www.biorxiv.org/content/10.1...
11.11.2025 17:14 — 👍 102 🔁 48 💬 3 📌 2
The constant barrage of terrible news on bluesky has made me feel weird about promoting papers, but people in the lab have been doing so much amazing work over the past few months that I want to share a few brief teasers/links:
10.09.2025 16:46 — 👍 66 🔁 22 💬 2 📌 1
Oh very cool—I think I missed that paragraph in your plos bio paper! Super interesting that these effects seem to matter in both yeast and E. coli
22.08.2025 15:17 — 👍 0 🔁 0 💬 0 📌 0

Frequency-dependent fitness effects are ubiquitous
In simple microbial populations, the fitness effects of most selected mutations are generally taken to be constant, independent of genotype frequency. This assumption underpins predictions about evolutionary dynamics, epistatic interactions, and the maintenance of genetic diversity in populations. Here, we systematically test this assumption using beneficial mutations from early generations of the Escherichia coli Long-Term Evolution Experiment (LTEE). Using flow cytometry-based competition assays, we find that frequency-dependent fitness effects are the norm rather than the exception, occurring in approximately 80\% of strain pairs tested. Most competitions exhibit negative frequency-dependence, where fitness advantages decline as mutant frequency increases. Furthermore, we demonstrate that the strength of frequency-dependence is predictable from invasion fitness measurements, with invasion fitness explaining approximately half of the biological variation in frequency-dependent slopes. Additionally, we observe violations of fitness transitivity in several strain combinations, indicating that competitive relationships cannot always be predicted from fitness relative to a single reference strain alone. Through high-resolution measurements of within-growth cycle dynamics, we show that simple resource competition explains a substantial portion of the frequency-dependence: when faster-growing genotypes dominate populations, they deplete shared resources more rapidly, reducing the time available for fitness differences to accumulate. Our results demonstrate that even in a simple model system designed to minimize ecological complexity, subtle ecological interactions between closely related genotypes create frequency-dependent selection that can fundamentally alter evolutionary dynamics. ### Competing Interest Statement The authors have declared no competing interest.
How common are frequency dependent fitness effects?
New preprint out today 👇
doi.org/10.1101/2025...
21.08.2025 19:23 — 👍 93 🔁 41 💬 6 📌 0

I'm very excited to share something I've been working on off-and-on for a long time now: a new blog about genotype-phenotype landscapes! The first post is a Gödel-Escher-Bach-style dialogue to introduce the topic. If you like it please share/repost! open.substack.com/pub/topossib...
27.07.2025 20:18 — 👍 11 🔁 5 💬 0 📌 0
New review article with @mmdesai.bsky.social is out today! Grateful for the opportunity to contribute something we hope will serve the community well
21.07.2025 17:30 — 👍 47 🔁 15 💬 3 📌 0
1/n 🧵 Excited to share our new paper! We developed a framework to reveal hidden simplicity in how organisms adapt to different environments, particularly focusing on antibiotic resistance evolution. #EvolutionaryBiology #MachineLearning
15.05.2025 14:33 — 👍 38 🔁 22 💬 1 📌 1

After a long and winding odyssey, excited to finally drop anchor in open-access waters. This preprint shows how neutral allele frequency time series can illuminate disease transmission rates between communities— key for epidemic fore- & backcasting. medrxiv.org/content/10.1... 🧵
06.12.2024 22:50 — 👍 29 🔁 11 💬 1 📌 2
I view genetic drift and decoupling noise as more fundamental demographic stochastic forces, which go on to affect downstream and emergent dynamics.
22.11.2024 17:50 — 👍 2 🔁 0 💬 0 📌 0
I think that is fair to say in one sense. The distinction I want to make is that genetic draft is emergent from an interplay of mutation, selection, etc. Changing population genetic parameters, including the strength of drift or decoupling noise, would also change genetic draft.
22.11.2024 17:49 — 👍 1 🔁 0 💬 1 📌 0
Thread from the preprint 👇
bsky.app/profile/joao...
22.11.2024 15:46 — 👍 0 🔁 0 💬 0 📌 0

Asynchronous abundance fluctuations can drive giant genotype frequency fluctuations
Nature Ecology & Evolution - Based on a combination of experiments and modelling, this study shows large stochastic fluctuations in genotype frequencies caused by intrinsic and extrinsic...
We usually think of genetic drift as the predominant stochastic force in evolving populations. But working with some model microbial populations, we found a distinct source of demographic stochasticity that scales (and behaves) differently than drift
Learn more in our new paper 👉 rdcu.be/d07Np
22.11.2024 15:46 — 👍 28 🔁 10 💬 2 📌 1
Yeah, genetic drift is dominant source of fluctuations at low frequencies, but then decoupling noise starts to dominate above frequencies ~1/(δ*N_e). So depending on the parameters, that cross-over point can be at a really low frequency. I don't know about recombination though, great question!
01.03.2024 06:58 — 👍 1 🔁 0 💬 0 📌 0
If you’ve gotten this far, thanks for reading and we welcome any feedback that you might have!
28.02.2024 22:39 — 👍 0 🔁 0 💬 1 📌 0
We spend a lot of time trying to measure fitness effects in evolution experiments, but comparatively little effort measuring the noise. I think that it is time to pay more attention to the fluctuations!
28.02.2024 22:39 — 👍 0 🔁 0 💬 1 📌 0
When we think of evolutionarily-important stochasticity, we usually think of genetic drift. But decoupling noise is like the shy cousin of drift—largely overlooked, but an important and likely common source of randomness in the frequencies of closely related genotypes.
28.02.2024 22:39 — 👍 0 🔁 0 💬 1 📌 0
Finally, we develop some new popgen theory. Some key findings: (1) Decoupling noise can significantly shift the ability of natural selection to distinguish between different fitness effects (2) Decoupling noise can leave selection-like signatures in the SFS
28.02.2024 22:38 — 👍 0 🔁 0 💬 1 📌 0

Because N^2-scaling abundance fluctuations are common across populations, we also think that decoupling noise may be ubiquitous. For example, we also find signatures of decoupling noise in the barcoded yeast experiments from the Petrov and Sherlock lab
28.02.2024 22:38 — 👍 0 🔁 0 💬 1 📌 0
The characteristic (Lyapunov) time is pretty fast—about 5-10 hours. So the dynamics look effectively stochastic if we’re taking samples every 24 hours. Only with these densely sampled time courses can we see the chaos.
28.02.2024 22:37 — 👍 0 🔁 0 💬 1 📌 0

So what is the cause of these fluctuations?
We cultured replicates and tracked the populations over a 24 hour cycle. The replicates exponentially diverge from each other! This is the signature of chaotic dynamics—small differences between replicates are exponentially amplified
28.02.2024 22:37 — 👍 0 🔁 0 💬 1 📌 0
Large frequency fluctuations may not be surprising if we were in a noisy environment. But we’re trying as hard as possible to maintain a constant environment, using closely related genotypes!
28.02.2024 22:37 — 👍 0 🔁 0 💬 1 📌 0
This is similar to previous models that invoke a fluctuating environment, but we know that many other mechanisms can cause these types of abundance fluctuations (e.g. chaos, aggregation, etc.)
28.02.2024 22:35 — 👍 0 🔁 0 💬 1 📌 0
But f^2-scaling frequency fluctuations don’t arise unless the abundance fluctuations are decoupled (to some degree) between the genotypes in the population. So we call these frequency fluctuations “decoupling noise”.
28.02.2024 22:35 — 👍 0 🔁 0 💬 1 📌 0

How do we explain this?
We developed a flexible model that can account for the scaling behaviors. Uncorrelated offspring number fluctuations causes classical genetic drift. In contrast, correlated offspring number fluctuations cause ~N^2-scaling abundance fluctuations.
28.02.2024 22:35 — 👍 0 🔁 0 💬 1 📌 0

Under classical genetic drift, the frequency variance should scale linearly with the mean. Instead, we saw a power-law relationship, with the variance scaling like the mean squared.
28.02.2024 22:35 — 👍 0 🔁 0 💬 1 📌 0
We initially didn’t know how the frequency fluctuations could be generated, and cycled through a couple different hypotheses. But then we performed a key experiment to measure the genotype frequency mean-variance scaling behavior.
28.02.2024 22:34 — 👍 0 🔁 0 💬 1 📌 0
Indeed, the fluctuations of neutrally barcoded subclones within each genotype is largely consistent with bottlenecking. So what is going on here?
28.02.2024 22:34 — 👍 0 🔁 0 💬 1 📌 0

It all started with our barcoded timecourses of a simple microbial population—we noticed huge frequency fluctuations. These populations live in a daily dilution culture, so we thought bottlenecking should drive noise. But the noise was orders of magnitude larger than expected!
28.02.2024 22:33 — 👍 0 🔁 0 💬 1 📌 0
Prof & Canada Excellence Research Chair | UBC Zoology, M&I | Oxford Biology | Evolution, ecology, host-pathogen, biodiversity | #firstgen | she/her/hers
Evolutionary systems/cell biologist. EMBO and SNSF Postdoctoral Fellow with Dmitri Petrov and Dan Jarosz at Stanford. PhD with Andreas Wagner at the University of Zurich. Studying how molecular and cellular systems shape, and are shaped by, evolution.
Assistant Prof at McGill, Oncology & Biochem departments, Lady Davis Institute | chromatin/txn | previous member @adelmanlab, LeAnn Howe labs | he/him | Fragile Nucleosome co-organizer | FHC MNT alumn 🏑🇨🇦🦌 | https://www.martinlab.ca/
The scientist formerly known as Lab Girl.
I write books: https://www.penguinrandomhouse.com/authors/2111914/hope-jahren/
Postdoc at UPenn thinking about mutations, cells, and evolution.
Publishing reviews and commentaries across the fields of genetics and genomics. Part of Springer Nature and Nature Portfolio.
https://www.nature.com/nrg/
Physics and biology of microbial communities. Structure, function, evolution. Center for the Physics of Evolving Systems. Dept. of Ecology and Evolution. UChicago. @NITMB, @CLS. kuehnlab.org
Biophysics PhD candidate and SF resident, studying models of microbial evolution and ecology with Ben Good at Stanford
https://tinyurl.com/jdmcenany
Assistant Professor, University of Washington, Genome Sciences.
Previous: JSMF Fellow, Berkeley EECS
♡: Computational biology, evolutionary dynamics, quantitative immunology
https://dewitt-lab.github.io/
[disclaimer: opinions mine]
Assistant Professor at UW Milwaukee | Molecular Microbiology and Evolution Lab | Microbes, Mentorship, and More | she/hers
Assistant prof at University of Rochester engineering genetic communication in microbial populations. Mountain enthusiast, passionate teacher
Postdoctoral researcher at Georgia State University
Microbiology, ecology, and evolution
Azimi Lab
Staff scientist @ Fred Hutch.
PhD Candidate | Evolution/Complex Systems, University of Michigan | kumawatb.com
any pronouns | Postdoc in the Margolis Lab at St. Jude | UAB Microbiology PhD | LSU alum | cat and plant parent | Views are my own | #NativeinSTEM #LGBTQinSTEM
Research scholar at Georgia Institute of Technology. Eco-evo | Host-pathogen interactions | Survival | Coexistence. Loves microbes and math. Self-taught illustrator. Frog enthusiast. Punk.
www.canankarakoc.com
Microbiologist 🧫🧪 | Microbe-Microbe interactions in infection | Mom of 👧🏼👦🏼🐶| Asst. Professor at UTK | Views my own
Postdoc in the Turner lab @ Yale EEB | chernandezsci.com