Links:
🔗 Paper: www.biorxiv.org/content/10.1...
💻 Code: github.com/MarksLab-Das...
9/9
Pascal Notin
@pascalnotin.bsky.social
Research in AI for Protein Design @Harvard | Prev. CS PhD @UniofOxford, Maths & Physics @Polytechnique
@pascalnotin.bsky.social
Research in AI for Protein Design @Harvard | Prev. CS PhD @UniofOxford, Maths & Physics @Polytechnique
Links:
🔗 Paper: www.biorxiv.org/content/10.1...
💻 Code: github.com/MarksLab-Das...
9/9
Congratulations to the entire RNAGym team @rohitarorayyc.bsky.social @murfalo.bsky.social @christianchoe.bsky.social @cshearer.bsky.social Aaron Kollasch, Fiona Qu, Ruben Weitzman, Artem Gazizov, @sarahgurev.bsky.social Erik Xie @deboramarks.bsky.social
8/9
The moderate performance across all tasks reveals exciting opportunities! Key directions: RNA-specific training data, integrating structure-function relationships, and improving non-canonical base pair prediction. RNAGym provides the standardized foundation for progress.
7/9
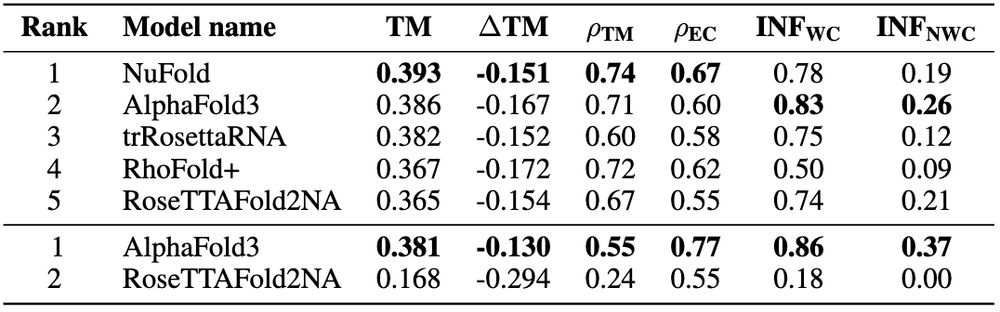
🌀 Tertiary structure: 215 diverse 3D structures from the PDB. NuFold leads monomers (0.393 TM-score), AlphaFold3 dominates complexes (0.381 TM-score). Non-Watson-Crick interactions remain a major challenge for all methods
6/9
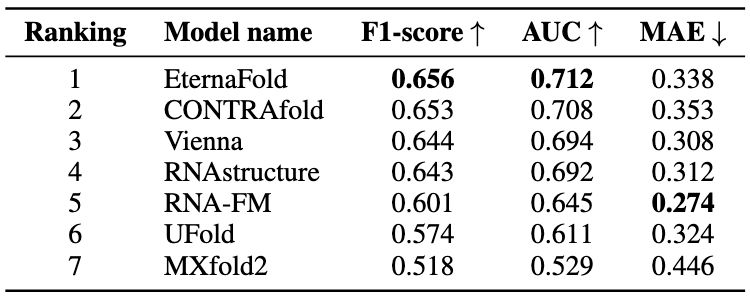
🔗 Secondary structure: 901k chemical mapping profiles using DMS & 2A3 reactivity. EternaFold achieves top performance (0.656 F1-score), closely followed by CONTRAfold & Vienna. Traditional thermodynamic methods are still competitive with newer deep learning approaches
5/9
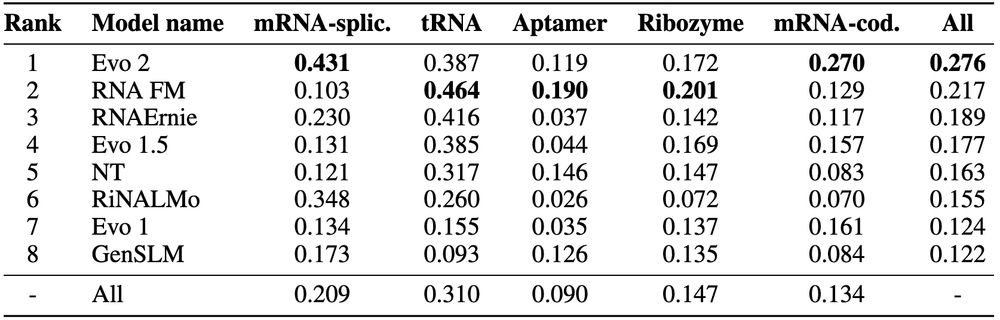
🔬 Fitness prediction: 70 assays across tRNA, ribozymes, aptamers & mRNAs (1M+ mutations total). Evo 2 performs best overall (0.276), but performance varies dramatically by RNA type: RNA-FM excels at tRNA/aptamers while Evo 2 leads mRNA tasks. Lots of room for improvement across the board!
4/9
RNAGym tackles three essential RNA prediction tasks: 🔬 Fitness prediction: How mutations affect RNA function 🔗 Secondary structure: Base-pairing patterns 🌀 Tertiary structure: 3D molecular architecture
All evaluated zero-shot to test true generalization!
3/9
Why do we need this? RNA modeling faces major challenges: limited experimental data (<1% of PDB entries), inherently less stable structures than proteins, and evaluation has been scattered across different studies with varying approaches.
2/9
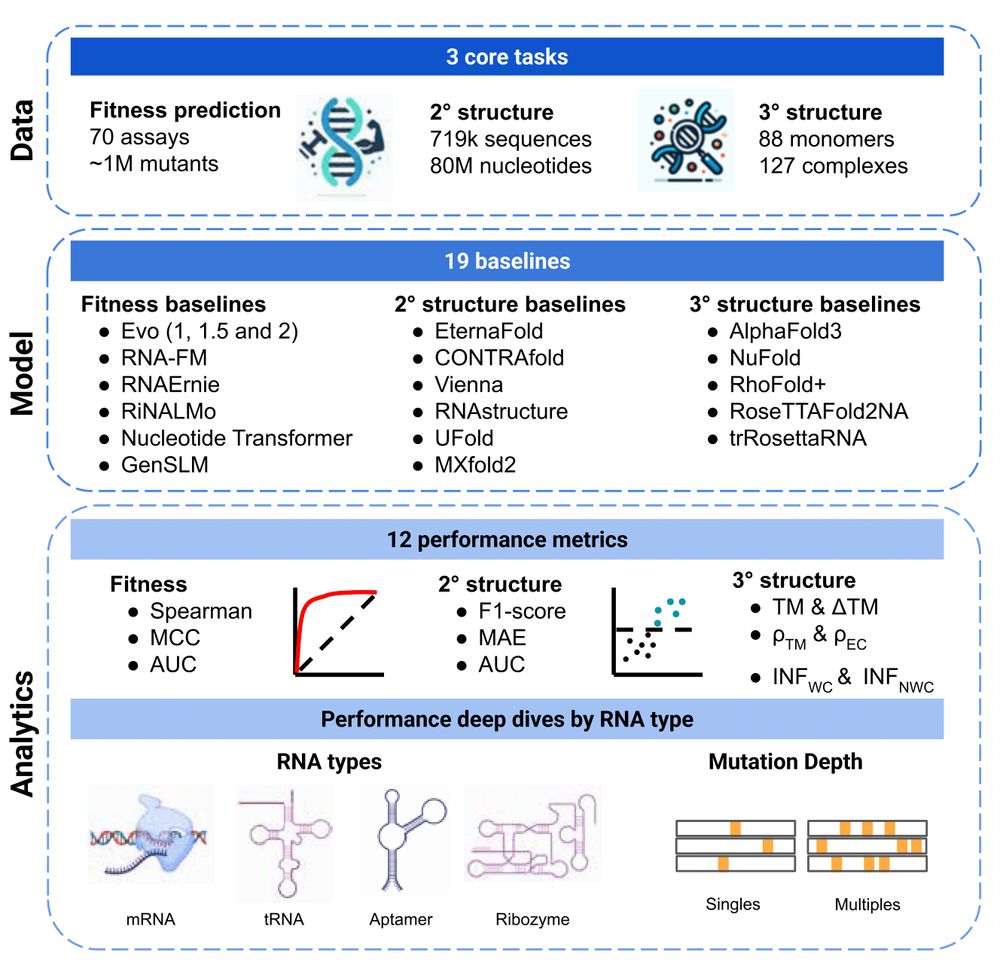
🚨 New paper 🚨 RNA modeling just got its own Gym! 🏋️ Introducing RNAGym, large-scale benchmarks for RNA fitness and structure prediction.
🧵 1/9
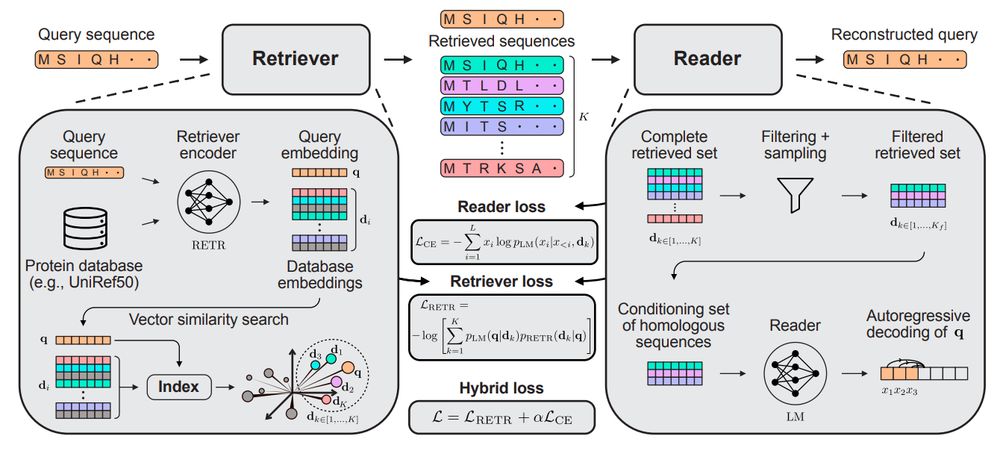
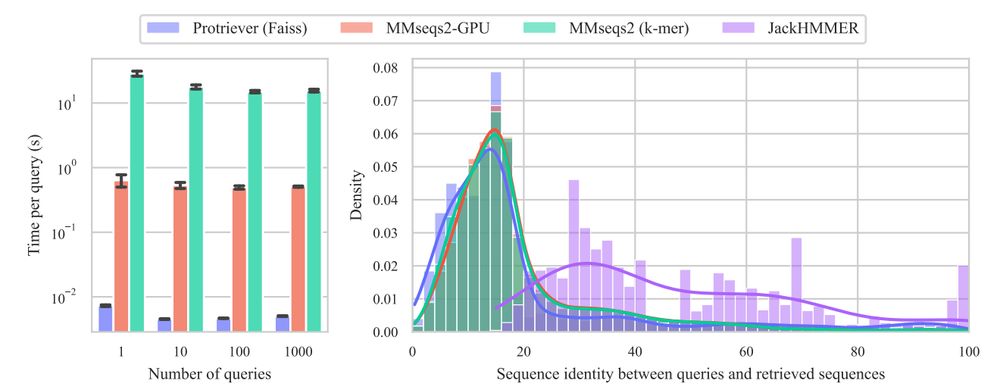
End-to-end differentiable homology search for protein fitness prediction.
@yaringal.bsky.social @deboramarks.bsky.social @pascalnotin.bsky.social
arxiv.org/abs/2506.089...
Pascal Notin at #VariantEffect25
21.05.2025 09:27 — 👍 11 🔁 2 💬 1 📌 0But more broadly I wanted to convey in the blog that the two (structure + MSA) are critical for proper functional protein design & effects prediction
08.05.2025 14:25 — 👍 2 🔁 0 💬 0 📌 0Thank you @delalamo.xyz! Understand where you are coming from re: design. For some design setups structure is critical -- here my point was more for a directed evolution setup where you have to select top mutants that go in the next round
08.05.2025 14:24 — 👍 1 🔁 0 💬 1 📌 0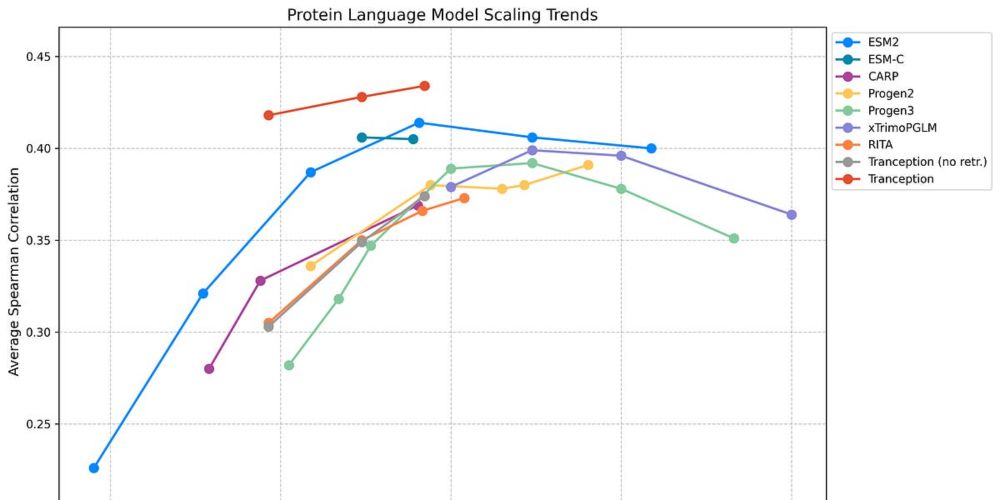
Even simple methods leveraging these 2 modalities significantly outperform billion-parameter sequence-only models. So, what's next? Better retrieval, advanced multimodal approaches, & alignment. Read more: pascalnotin.substack.com/p/have-we-hi... #BioTech #AI #pLMs
08.05.2025 00:29 — 👍 8 🔁 1 💬 1 📌 1Have we hit a "scaling wall" for protein language models? 🤔 Our latest ProteinGym v1.3 release suggests that for zero-shot fitness prediction, simply making pLMs bigger isn't better beyond 1-4B parameters. The winning strategy? Combining MSAs & structure in multimodal models!
08.05.2025 00:29 — 👍 25 🔁 7 💬 1 📌 2Large-scale discovery, analysis, and design of protein energy landscapes https://www.biorxiv.org/content/10.1101/2025.03.20.644235v1
25.03.2025 14:47 — 👍 10 🔁 8 💬 0 📌 1