
NSF Invests Nearly $32M to Accelerate Novel AI-Driven Approaches in Protein Design
ow.ly/NJzp50WCc00 #NSF #AIwire
@fabienplisson.bsky.social
BioDesign, Machine Learning, Drug Discovery | Rosenkranz Award 2021 | Dad | Polyglot | Capybarist | plissonf.github.io Founding ingeniebio.com ORCID 0000-0003-224

NSF Invests Nearly $32M to Accelerate Novel AI-Driven Approaches in Protein Design
ow.ly/NJzp50WCc00 #NSF #AIwire
Worth a watch:
Head of Signal, Meredith Whittaker, on so-called "agentic AI" and the difference between how it's described in the marketing and what access and control it would actually require to work as advertised.
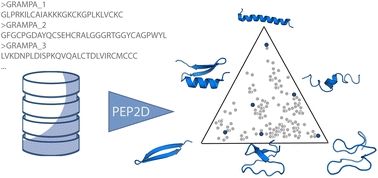
If that helps, we compared AF2 predicted structures of a small set of peptides against their PDB structures under different experimental methods (NMR solutions, X-ray, SSNMR, EM) as ground truth. pubs.rsc.org/en/content/a...
28.06.2025 08:25 — 👍 2 🔁 0 💬 0 📌 0Think coronavirus spikes have run out of surprises? Think again.
Our latest preprint dives into the highly unusual spikes of marine mammal coronaviruses.
www.biorxiv.org/content/10.1...
This #cryoEM study was led by @viralfusion.bsky.social, with key contributions from an amazing team.
Derguini, F.; Plisson, F.; Massiot, G. 𝘗𝘳𝘦𝘱𝘢𝘳𝘢𝘵𝘪𝘰𝘯 𝘰𝘧 𝘵𝘢𝘨𝘪𝘵𝘪𝘯𝘪𝘯 𝘊 𝘢𝘯𝘥 𝘍 𝘥𝘦𝘳𝘪𝘷𝘢𝘵𝘪𝘷𝘦𝘴 𝘢𝘴 𝘢𝘯𝘵𝘪-𝘤𝘢𝘯𝘤𝘦𝘳 𝘢𝘨𝘦𝘯𝘵𝘴. FR 2941697 A1 20100806 2010.
Europe PMC europepmc.org/article/PAT/...
Google Patents
patents.google.com/patent/FR294...
As an engineer at heart, I wondered how we would scale up if we found a non-toxic, bioactive analogue. That led us to 𝗯𝗶𝗼𝗰𝗮𝘁𝗮𝗹𝘆𝘀𝗶𝘀 (think immobilised enzymes) - screening lipases and esterases to reach, in 1 step, a key intermediate: Taginitol C. This work was patented.
05.05.2025 01:32 — 👍 0 🔁 0 💬 1 📌 0Over three years, we synthesised dozens of Taginitin C analogues, eventually streamlining the synthesis from 10 to 6 steps 𝘷𝘪𝘢 judicious protection/deprotection strategies.
05.05.2025 01:32 — 👍 0 🔁 0 💬 1 📌 0The molecule was notoriously sensitive to acids, bases, nucleophiles, and UV light. Resources were limited, and we were performing multi-step syntheses on just 𝟱 𝗺𝗴 𝘀𝗰𝗮𝗹𝗲𝘀, in 𝘁𝗶𝗻𝘆 𝗿𝗼𝘂𝗻𝗱-𝗯𝗼𝘁𝘁𝗼𝗺 𝗳𝗹𝗮𝘀𝗸𝘀, working in darkened fume hoods.
05.05.2025 01:32 — 👍 0 🔁 0 💬 1 📌 0Together with various teammates (MSc students) we worked on the hit-lead optimisation of Tagitinin C - a toxic germanocrolide inhibiting the ubiquitin-proteasome pathway.
05.05.2025 01:32 — 👍 0 🔁 0 💬 1 📌 0Eighteen years ago, I joined one of my first drug development R&D programs at the 𝘐𝘯𝘴𝘵𝘪𝘵𝘶𝘵 𝘥𝘦𝘴 𝘚𝘤𝘪𝘦𝘯𝘤𝘦𝘴 𝘦𝘵 𝘛𝘦𝘤𝘩𝘯𝘰𝘭𝘰𝘨𝘪𝘦𝘴 𝘥𝘶 𝘔𝘦𝘥𝘪𝘤𝘢𝘮𝘦𝘯𝘵 𝘥𝘦 𝘛𝘰𝘶𝘭𝘰𝘶𝘴𝘦 - a public-private partnership between the @cnrs.fr and Pierre Fabre Laboratories.
05.05.2025 01:32 — 👍 0 🔁 0 💬 1 📌 0
Never too late to publish, I am proud to see this work finally published: Tagitinin C, a Sesquiterpene Lactone, and Derivatives as Proteasome Inhibitors doi.org/10.1002/ejoc...
05.05.2025 01:32 — 👍 1 🔁 0 💬 1 📌 0
Choosing ML architectures for protein engineering is often challenging. Our “new” updated preprint provides a rational framework to match ML models to protein fitness tasks, showing landscape ruggedness influences prediction accuracy. Mahakaran dana Adam et al www.biorxiv.org/content/10.1...
22.04.2025 12:41 — 👍 14 🔁 3 💬 0 📌 1
Run BioEmu in Colab - just click "Runtime → Run all"! Our notebook uses ColabFold to generate MSAs, BioEmu to predict trajectories, and Foldseek to cluster conformations.
Thanks @jjimenezluna.bsky.social for the help!
🌐 colab.research.google.com/github/sokry...
📄 www.biorxiv.org/content/10.1...
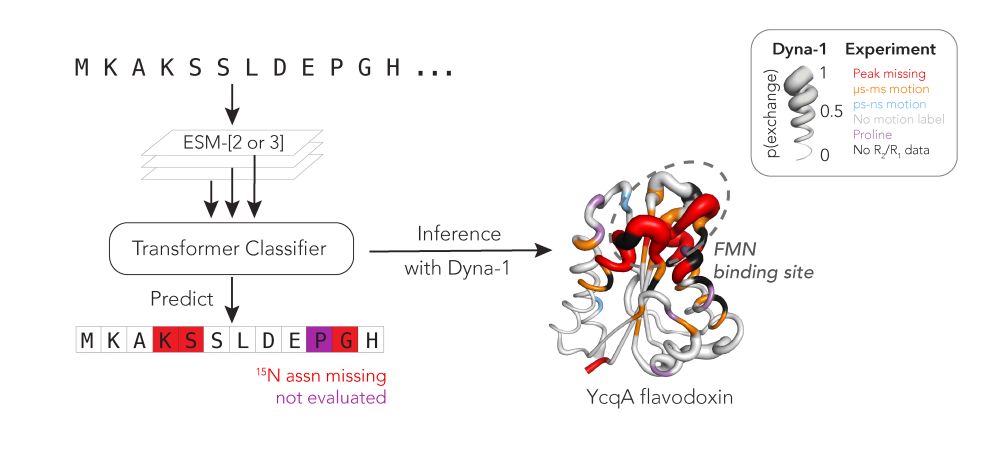
Protein function often depends on protein dynamics. To design proteins that function like natural ones, how do we predict their dynamics?
@hkws.bsky.social and I are thrilled to share the first big, experimental datasets on protein dynamics and our new model: Dyna-1!
🧵
You can download my protein structure-inspired artwork from pdb webpage:
pdb101.rcsb.org/sci-art/bezs...
@rcsbpdb.bsky.social
@pdbeurope.bsky.social
#sciart
YouTube is the world's 2nd-largest search engine. So why aren't more conference keynotes and presentations there? 🤔
19.02.2025 15:53 — 👍 7 🔁 3 💬 2 📌 0
Modern-Day Oracles or Bullshit Machines?
Jevin West (@jevinwest.bsky.social) and I have spent the last eight months developing the course on large language models (LLMs) that we think every college freshman needs to take.
thebullshitmachines.com
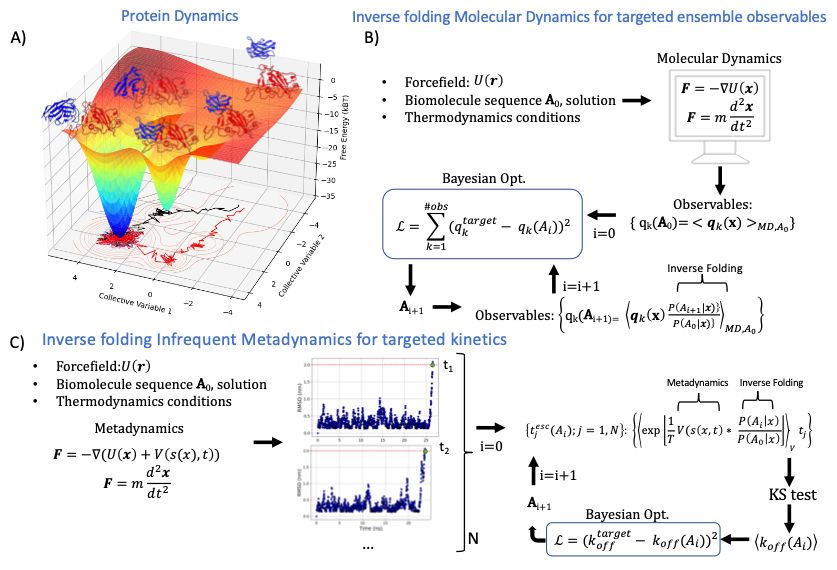
Fig. 1 Inverse Folding Molecular Dynamics (IF-MD) protocol. (A) Free energy landscape of the process of protein-protein dissociation in the case of SARS-CoV-2 RBD (red) in complex with the nanobody H11 (blue) visiting different metastable states along the unbinding mechanism along two collective variables. Two different unbinding trajectories from the bound state are shown in black and red. (B) IF-MD protocol to sample protein sequence space by constraining k ensemble averaged observables q target k and using Bayesian Optimization to propose new sequences. (C) Specific example of IF-MD constrained to a target the unbinding rate constant, and using Infrequent Metadynamics to sample the prior ensemble. Unbinding example trajectories sampled by Infrequent Metadynamics are shon in blue, along the RMSD from the cryo-EM structure of H11 bound to SARS-CoV-2 RBD as a function of time.
Short thread about this interesting preprint that explores antibody design by combining MD with inverse folding and active learning. It's a bit rough around the edges but it introduces a cool idea I hope is further fleshed out www.biorxiv.org/content/10.1...
17.02.2025 08:14 — 👍 35 🔁 6 💬 1 📌 0Are protein language models the universal key?
doi.org/10.1016/j.sb...
Brilliant and thoughtful piece.
Brilliant video on the development of protein structure prediction featuring #AF2 and #Rosetta series
youtu.be/P_fHJIYENdI?...
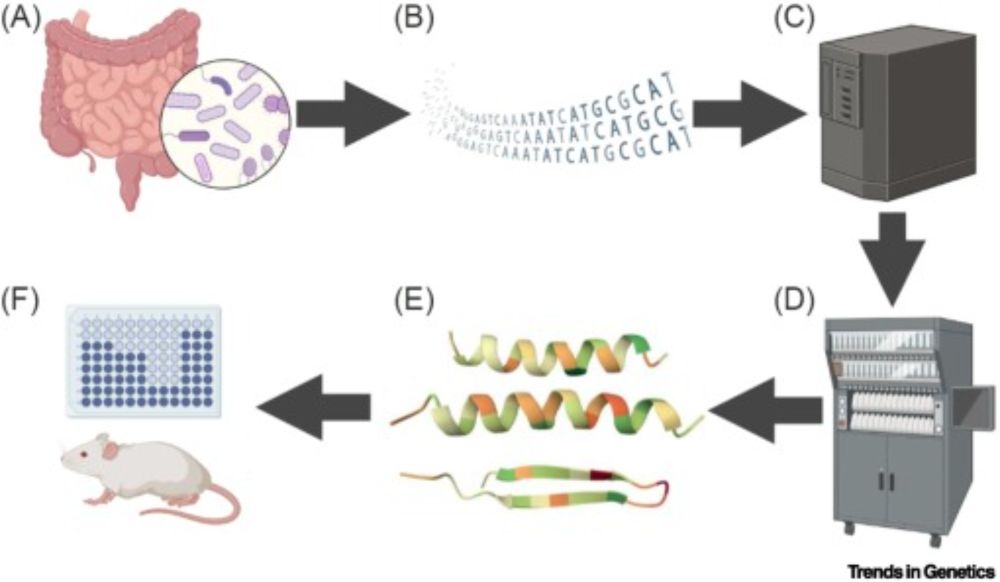
(1/4)In our new paper in @cp-trendsgenetics.bsky.social @cellpress.bsky.social, we highlight some fascinating tiny proteins—microproteins—which appear to be widespread in nature. www.cell.com/trends/genet...
05.02.2025 14:00 — 👍 8 🔁 4 💬 1 📌 0
Great editorial from @evotec.bsky.social using AI+ML tools to assist drug discovery campaigns in the small and middle chemical spaces
pubs.acs.org/doi/10.1021/...
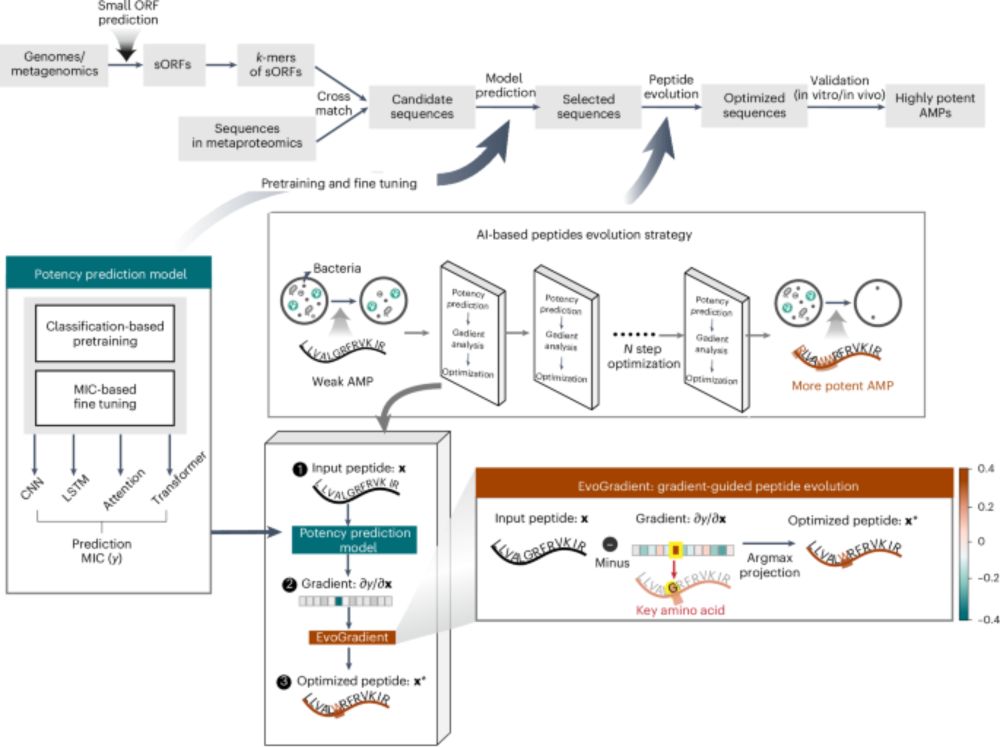
Beilun Wang and team recently reported the development of antimicrobial peptides combining an ensemble of DL predictors with EvoGradient, modifying candidate sequences strategically from evolutionary information, improving their antimicrobial activity and selectivity.
www.nature.com/articles/s41...
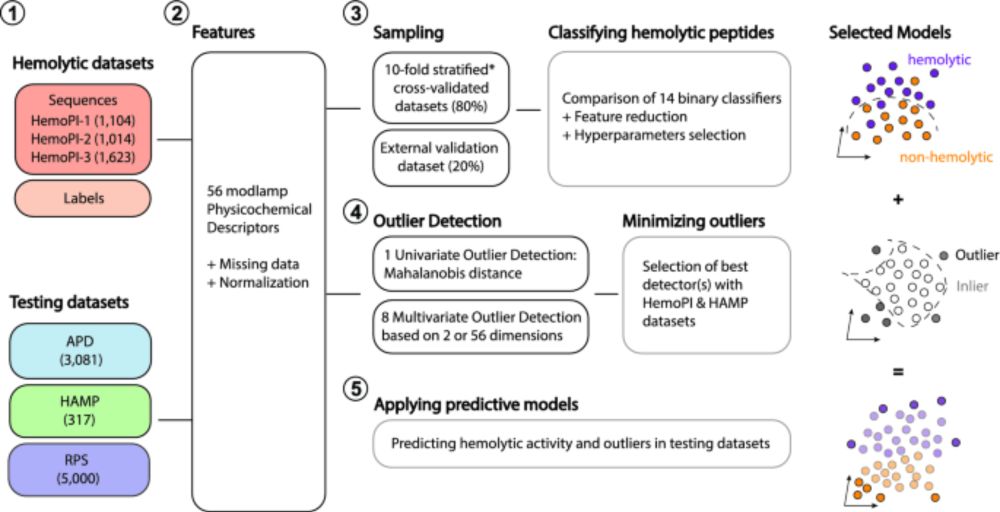
We previously used that strategy to set guidelines for discovering and designing non-hemolytic peptides:
www.nature.com/articles/s41...
The process mimics the hypothesis-driven process of a medicinal chemist, especially with Deep Mutational Scanning. It also merits explaining the results from your predictive model(s) by comparing mutants.
29.01.2025 23:37 — 👍 0 🔁 0 💬 1 📌 0Another strategy starts from sequences identified with the best predictive scores and applies a generative algorithm to create analogues (via random mutagenesis or directed evolution), followed by predictions.
29.01.2025 23:37 — 👍 0 🔁 0 💬 1 📌 0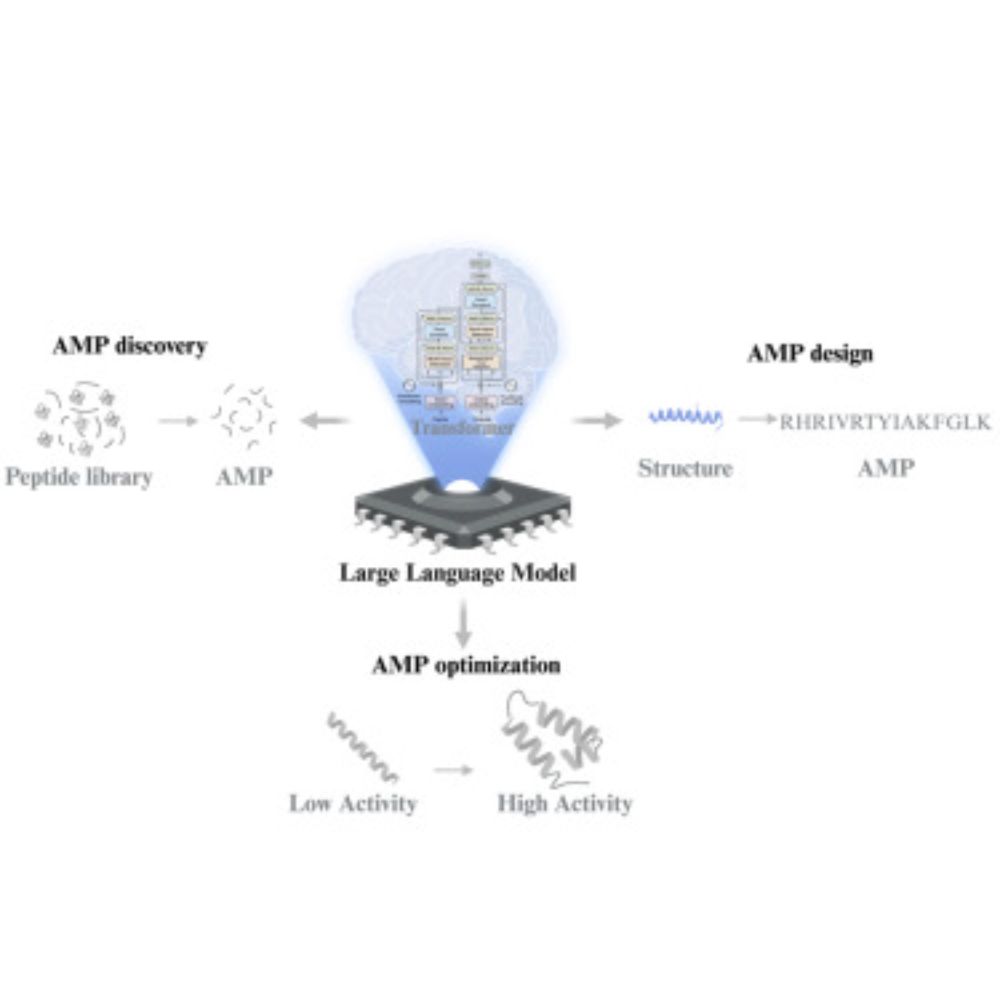
A common strategy for identifying sequences with the desired characteristics is to combine a generative algorithm with multiple models predicting properties and functions. This narrows the search in the vast peptide fitness space.
www.sciencedirect.com/science/arti...

Generative models have created peptide sequences that share antimicrobial characteristics (e.g., amphipathic character, charged residues, helical constraint).
link.springer.com/protocol/10....
These models are routinely used to discover novel AMPs from biodiverse extant and extinct species. Check out the great work done by @delafuentelab.bsky.social team with their APEX models.
29.01.2025 23:37 — 👍 1 🔁 0 💬 1 📌 0Closer to my experience in #CompBiology - the development of AI algorithms to discover and design antimicrobial peptides against #AMR, where predictive ML models are employed to predict the antimicrobial nature (classification) or activity (regression), primarily from sequences.
29.01.2025 23:37 — 👍 1 🔁 0 💬 1 📌 0