Exciting new #Zebrafish research from the #WeinsteinLab, led by Jong Park!
“Specialized gas-exchange endothelium of the zebrafish gill” —
www.biorxiv.org/content/10.6...
Amazing to see red blood cells moving through the gills! Don’t forget to check out the supplemental movies ;-)
02.12.2025 22:48 — 👍 2232 🔁 328 💬 36 📌 21
Lerche, Mathieu, @johannaivaska.bsky.social @ivaskalab.bsky.social et al. reveal that newly synthesized #integrins can bypass the Golgi to be actively delivered to specific plasma membrane regions, where they enhance cell protrusion and #adhesion. rupress.org/jcb/article/...
02.12.2025 18:30 — 👍 29 🔁 12 💬 0 📌 0
🤩🤩🤩
25.11.2025 20:52 — 👍 1 🔁 0 💬 0 📌 0

It mourns not for its own fate, but for that of the world and all its beauty, flitting through the cosmos, blown into the void like a light blue wildflower trampled by the night's frost.
06.11.2025 16:59 — 👍 76 🔁 15 💬 11 📌 1
Expansion Microscopy keeps getting better!
01.11.2025 18:27 — 👍 5 🔁 0 💬 0 📌 0
In the spirit 👻 of Freaky Friday 🎃, here is a beautiful yet eery video of zebrafish retina development showing horizontal cells (🔵) finding their way out of the crowded amacrine (🟠) layer to settle beneath the photoreceptors. Happy #Halloween!
🎥: PhD student Rae Wong from the @nordenlab.bsky.social
31.10.2025 14:29 — 👍 42 🔁 18 💬 0 📌 0
Apply - Interfolio
{{$ctrl.$state.data.pageTitle}} - Apply - Interfolio
During these uncertain times, I’m very happy to see that my institution, @scripps.edu has an open tenure-track Assistant Professor position. Any field in Chemistry or Biology is welcome. I’d especially love to see fellow neuroscientists apply. Please repost!
apply.interfolio.com/174756
14.10.2025 17:35 — 👍 130 🔁 95 💬 2 📌 5

We hire!
The CytoMorpho Lab is looking for an engineer with a background in cell and molecular biology to join our team in Paris. This is a 18-month contract position, but we're open to exploring long-term opportunities.
More details here:
cytomorpholab.com/index.php/jo...
14.10.2025 13:50 — 👍 82 🔁 73 💬 0 📌 0
I found these immune cells just finished mitosis but their midbody is still yet to be resolved. How cool? If you zoom in to the midbody, you can see its classical shape just like in the textbooks. I love these little details.
14.10.2025 10:55 — 👍 113 🔁 28 💬 4 📌 1
The future of High-Content Imaging has arrived.
-> Ultra-fast 3D imaging of multi-well plates at high resolution using an air objective!
See previous posts and the new miOPM preprint for details: doi.org/10.1101/2025...
13.10.2025 00:44 — 👍 39 🔁 9 💬 2 📌 1
Playing around with how to represent volume in #Snouty light sheet data. Here is a dividing hiPSC with endogenously tagged histone in all three orthogonal projections. Bluer is further away. If anything, it distracts from politics.
08.10.2025 19:57 — 👍 32 🔁 9 💬 1 📌 0
🍀🔬
MSL10 is a high-sensitivity mechanosensor in the tactile sense of the Venus flytrap @natcomms.nature.com from Toyota lab.
www.nature.com/articles/s41...
01.10.2025 14:57 — 👍 24 🔁 7 💬 0 📌 1
SCHEPHERD--the bioelectric cell herding platform built for YOU. Single cells, monolayers, organoids--this herds them all + new tricks. Plz try it-- we will *give* you parts! Teaser here of a steering a single cell. GS Yubin Lin's lifeblood with J. Yodh on piano; Celeste R. and Paul K. Thread 1/N
17.09.2025 17:20 — 👍 61 🔁 26 💬 4 📌 4

My group at Science has been running a big campaign to engineer new channelrhodopsins.
Excited to share the first results from it today: a suite of highly sensitive new opsins.
We call them "WAChRs".
Everyday indoor office lighting is enough to activate them pretty strongly.
18.09.2025 14:06 — 👍 48 🔁 10 💬 6 📌 2

E-cadherin mechanotransduction activates EGFR-ERK signaling in epithelial monolayers by inducing ADAM-mediated ligand shedding
Epithelial stretching promotes the release of EGF receptor ligands that stimulate ERK activation.
A bit delayed, but excited to still share our latest paper, showing that intercellular forces transduced by E-cadherin activate EGFR-ERK signaling in epithelia by inducing EGFR ligand shedding! Mechanical and biochemical signals can act together within a single, linear cascade! tinyurl.com/mr9mj9j2
13.09.2025 18:04 — 👍 69 🔁 25 💬 4 📌 0
Ant reproduction is getting weirder and weirder. Here’s the latest: queens of a Mediterranean harvester ant mate with males from a different species and then lay eggs that clonally inherit those males’ genome. I.e., they lay eggs that develop into a different species. 🤯
05.09.2025 22:07 — 👍 52 🔁 25 💬 1 📌 2
I’ve already highlighted this landmark #lipidtime paper by @nadlerlab.bsky.social & colleagues, but ICYMI, do read André’s thread — this study is a huge leap forward in understanding the logic of intracellular lipid flux & is a phenomenal example of #chembio probes 🤝 quantitative imaging 🤝 modeling!
31.08.2025 17:35 — 👍 25 🔁 5 💬 2 📌 0

Nice! A song about one of my favorite fluorescent proteins! mStayGold!
30.08.2025 11:23 — 👍 6 🔁 1 💬 0 📌 0
Congrats!
01.08.2025 21:13 — 👍 1 🔁 0 💬 0 📌 0
🤩
16.07.2025 07:25 — 👍 0 🔁 0 💬 0 📌 0
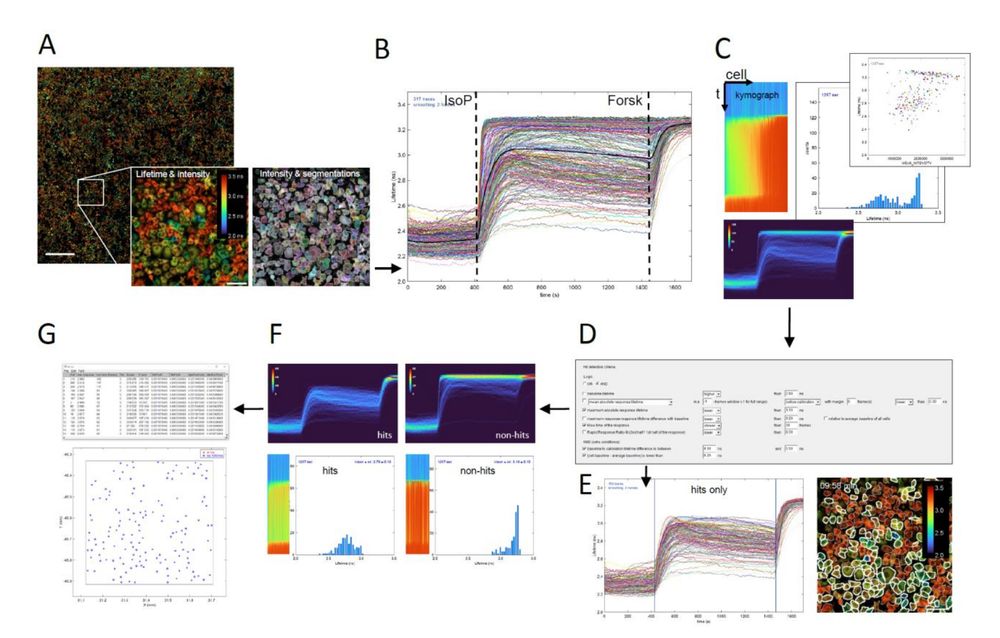
Fig. 4. Time-lapse FLIM analysis and hit detection Fiji macro. (A) Stitched multi-tile intensity image
overlay with the fluorescence lifetime image. The lifetime image can be either average photon arrival times
(‘Fast FLIM’) or calculated average lifetimes from two lifetime components. Ratiometric images can also
be used. Scale bar: 1000 μm. Inset: Left – Zoom-in of lifetime image of cells after stimulation with
isoproterenol; Right – Zoom-in showing cell segmentation as outlines. Scale bar: 100 μm. (B) Lifetime time
traces of all 298 segmented cells in a single tile of Cos7 cells (a mix of ADRB2 KO and WT cells). Cells are
stimulated with 40 nM isoproterenol (IsoP), followed by a calibration with forskolin (Forsk). Stimulation
and calibration time points may be automatically calculated from the average of all traces (thick black
line) or entered manually. (C) The script generates several informative visualizations. Shown are a
‘kymograph’ representation of fluorescence lifetime vs time in all cells, sorted on response magnitude (top-
left); a normalized data density graph (2D histogram of lifetime vs time, with cell counts in false color),
convenient when analysing thousands of cells (bottom); and (optionally animated) histograms of the
fluorescence lifetimes over time (middle) and scatterplots of lifetime vs secondary parameters like cell
intensity, cell area or intensity of an additional fluorescence channel of choice (top-right). (D) Hit selection
dialog showing various optional criteria. These can be used separately or combined, allowing to screen for
a large variety of dynamic phenotypes. (E) Based on the criteria, hit cells are determined and visualized
through a ‘hits only’ lifetime traces plot (left). Blue vertical lines indicate the chosen hit detection time
window. The hit cells are outlined with white lines on the lifetime-intensity overlay image (right). (F)
Additional visualizations of hit cells (left) and non-hit cells (right), in density graphs, resp…
Beyond Static Screens: A High-Throughput Pooled Imaging CRISPR Platform for Dynamic Phenotype Discovery by Kees Jalink and team: www.biorxiv.org/content/10.1...
14.07.2025 07:35 — 👍 15 🔁 3 💬 2 📌 0
Biggest real problem Europe has is that after a certain career age, it’s Advanced or bust for many people. And there’s VERY few ERC advanced grants (way lower number than R01s). This creates a huge problem for recruitment, especially from outside, which is supposedly what they’re trying to improve.
13.07.2025 09:58 — 👍 11 🔁 4 💬 1 📌 0
🤩
11.07.2025 06:42 — 👍 0 🔁 0 💬 0 📌 0
Cool idea that stems from integrating learnings from animal behaviorists with leading edge vaccine technology 👏
09.07.2025 15:24 — 👍 17 🔁 5 💬 0 📌 0
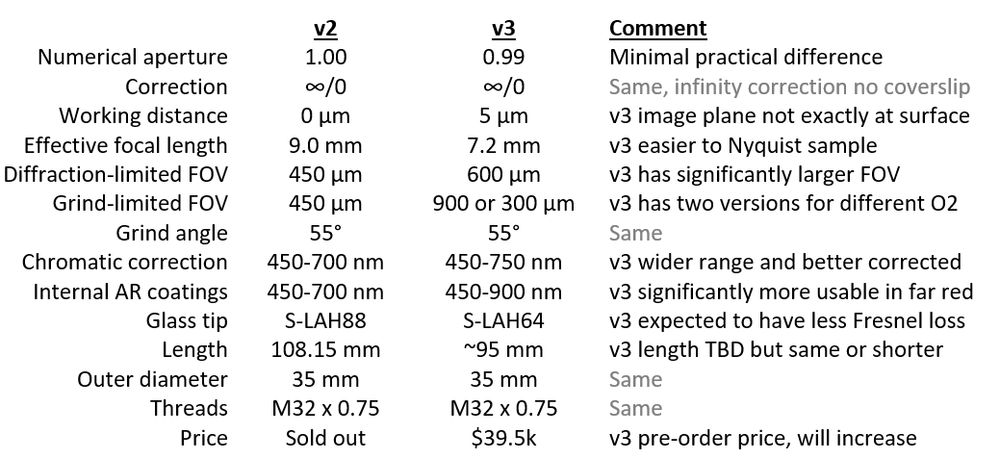
comparison table for v2 and v3
Production has started for a new #Snoutscope objective, AMS-AGY v3. It has significantly larger FOV and some other tweaks from v1 and v2. Currently accepting pre-orders. The price will need to increase modestly after deliveries start late October.
09.07.2025 20:00 — 👍 26 🔁 12 💬 4 📌 1

Possible End to End to End Encryption: Come Help - Bert Hubert
tl;dr: The European Commission is honestly asking for experts to advise them on ways to institute “effective and lawful access to data for law enforcement”. If you are an expert, I urge you to apply t...
Attempts by law enforcement & governments to subvert end-to-end encryption are ongoing. The European Commission will spend a year thinking about their new "Roadmap for law enforcement access to data" & they are (genuinely) asking for people to join an expert group to help: berthub.eu/articles/pos...
05.07.2025 11:43 — 👍 20 🔁 16 💬 4 📌 0
Using genomic information to improve medicine
🌱 Grow Food 🌎 Heal the Planet
🐝 For gardeners learning to grow regeneratively ♻️
📚 FREE knowledge + community on our website! ↙️
https://learndirt.com/
PhD Student @ Syracuse University
protists, extremophiles, & science art | she/her | 🔬🏳️🌈
🎙️ Live, from 19p13, it's Saturday Night!
https://en.wikipedia.org/wiki/LKB1
@JohanaProtists | Assistant Professor at Department of Marine Sciences University of Puerto Rico Mayagüez | MIC.Rott Lab | Protists | Symbioses in Marine Anaerobes | Microbial Interactions in Ciliates | www.microttlab.org | newPI
Group leader @mpi-mg, Berlin
Chromatin tracing, epigenetics, development. Equity and advocacy in academia.
Director, Dept. of Mechanistic Cell Biology, Max Planck Institute of Molecular Physiology, Dortmund (DE). Opinions my own
Organic Chemist & Biochemist | Chemical Biology | Triathlete
Not-for-profit research institute advancing the science of genomics through bold innovations to improve human health and environmental sustainability. Learn more at https://www.jcvi.org.
⚡ Extreme Weather | 🌪️ Storm Chaser | Meteoroloog Weerplaza (media & bewaking gladheid) | Natuurfotografie | Space Weather ☄️ | Persoonlijk account | www.woutervanbernebeek.nl
Historian of science & medicine and writer @ Johns Hopkins & Berkeley. Biography of James Watson coming soonish from Basic Books. Also rock climbing, roots music, tattoos, dogs, humor. My opinions are his –>
From embryos to equations and back.
We study how physical principles shape life, from collective DNA–protein interactions to mesoscale processes within cells and tissues.
Tweets by Grill Lab members.
https://grill-lab.org
Researching why legless lizards and snakes don't have feet. (She/her) 😷
convergent evolution, paleontology, ADHD, disability rights, grants, whatever my hyperfixation is this week.
kkoeller3115.wixsite.com/kristakoeller
https://twitter.com/KristaLerista
Cell migration and endocytosis research lab
https://cytoskeleton.wixsite.com/krauselab
The fluorescent protein database:
https://www.fpbase.org
Account by @talleylambert.bsky.social
Structural biologist, Cryo-EM aficionado, signal peptide nerd, teacher, mentor, dad, …
ISDB is a non-profit scientific association that promotes the study of developmental biology
(she/her) Scientist, Berlin-based, PhD Student in @ewerslab.bsky.social
Here for Microscopy, Cytoskeleton, Cellbio, Cytokinesis. Black in STEM
Postdoc at CRG Barcelona
Biochemistry | Cytoskeleton | Cell Development | In vitro reconstitutions


















