Checkout the latest preprint from the lab - single-cell CIN signatures unlocked! Ongoing HRD improves PARPi sensitivity detection and ongoing NHEJ associates with subclonal diversification in TNBC
28.11.2025 17:36 — 👍 7 🔁 4 💬 0 📌 0

Fluctuating DNA methylation tracks cancer evolution at clinical scale
Nature - Cancer evolutionary dynamics are quantitatively inferred using a method, EVOFLUx, applied to fluctuating DNA methylation.
Cancer is an evolutionary disease, but does knowing a cancer’s evolutionary past help predict its future? Out today in @nature, we learnt the evolution of 2000 lymphoid cancers and found it was highly correlated with clinical outcomes! (1/7)
rdcu.be/eFrrc
10.09.2025 16:17 — 👍 46 🔁 19 💬 1 📌 2
12/ Thanks to patients and funders for their support @cniostopcancer.bsky.social @isciiisalud.bsky.social isciii.bsky.social @cienciagob.bsky.social #BecariosFLC #illumina @innovateuk.bsky.social @cuh.nhs.uk #TailorBio @cruk-ci.bsky.social
23.06.2025 09:02 — 👍 1 🔁 0 💬 1 📌 0
11/ Kudos to @jsneaththompson.bsky.social, Laura Madrid, @bhernando.bsky.social & co-authors for driving this work!
23.06.2025 09:02 — 👍 1 🔁 0 💬 1 📌 0
10/ What’s next? We’re funded by @mintradigital.bsky.social #NextGenerationEU for analytical validation and will be ready to run prospective trials in 2026. From organoids to algorithms to patients: precision chemo is possible!
23.06.2025 09:02 — 👍 0 🔁 0 💬 2 📌 0

9/ To ensure a flexible pathway to the clinic, we also tested biomarker reproducibility in ctDNA samples and TSO500 panel data, bringing us closer to real-world implementation
23.06.2025 09:02 — 👍 0 🔁 0 💬 1 📌 0

8/ It worked! We emulated trials to validate resistance predictions to platins, taxanes & anthracyclines across ovarian, breast, prostate & sarcoma
23.06.2025 09:02 — 👍 0 🔁 0 💬 1 📌 0

7/ Next step? A prospective trial? We tried but couldn’t! No one wanted to run/fund a trial using “old” chemos. So we had to get creative. Luckily, as chemos are widely used, there was a wealth of real-world data eg TCGA & @HartwigMedical to emulate biomarker trials - even RCTs!
23.06.2025 09:02 — 👍 0 🔁 0 💬 1 📌 0

6/ Our results looked great!
23.06.2025 09:02 — 👍 0 🔁 0 💬 1 📌 0
5/ Next step? Proof-of-concept using retrospective data from 50 ovarian cancer samples. Ovarian cancers were ideal as all 3 chemotherapies are routinely used. We focused on predicting resistance. Why resistance? Because it allows patients to avoid toxic side-effects
23.06.2025 09:02 — 👍 0 🔁 0 💬 1 📌 0

4/ We focused on optimising 3 CIN signature-based biomarkers to classify patients as resistant or sensitive to 3 commonly used chemotherapies: platins, taxanes or anthracyclines. Our goal: to optimise biomarker thresholds to use pan-cancer
23.06.2025 09:02 — 👍 0 🔁 0 💬 1 📌 0
3/ These prelim data showed correlations between CIN signatures and chemotherapy response. As the full spectrum of CINsigs can be quantified in a tumour using a single genomic test, we hypothesised that CINsigs could predict resistance to multiple chemotherapies at diagnosis
23.06.2025 09:02 — 👍 0 🔁 0 💬 1 📌 0
2/ Back then, we already had preliminary data suggesting these CIN signatures may be useful as therapy response biomarkers, mainly via synthetic lethality with the mechanism of action of the drug (CIN signature➡️defective pathway➡️dependency, which the drug exploits)
23.06.2025 09:02 — 👍 1 🔁 0 💬 1 📌 0

A pan-cancer compendium of chromosomal instability - Nature
Copy number signatures characterize different types of chromosomal instability and predict drug response.
1/ 3 yrs ago we developed a computational framework to decode chromosomal instability www.nature.com/articles/s41...: input a tumour genome➡️output CIN signatures. As these CIN signatures represent different causes of DNA damage, they provide a read out of defective pathways in a tumour
23.06.2025 09:02 — 👍 0 🔁 0 💬 1 📌 0
🚨Chemo treatment upgrade!🚨
Check out our approach to modernise chemotherapy treatment published today in @natgenet.nature.com. From @cniostopcancer.bsky.social #TailorBio @cruk-ci.bsky.social www.nature.com/articles/s41... More details 👇
23.06.2025 09:02 — 👍 16 🔁 9 💬 1 📌 2
Bluesky
@blaschaves.bsky.social @mescobarrey.bsky.social @torresmarina.bsky.social
10.06.2025 14:15 — 👍 0 🔁 0 💬 0 📌 0
…and to the patients and funders @cniostopcancer.bsky.social @isciiisalud.bsky.social @cienciagob.bsky.social #BecariosFLC #H12O
10.06.2025 14:15 — 👍 0 🔁 0 💬 1 📌 0
We hope this study will inspire further forecasting efforts across other molecular alterations. Thanks to all members of the @gmaci.bsky.social Lab & collaborators Paz Ares Lab #PPCG 14/
10.06.2025 14:15 — 👍 1 🔁 0 💬 1 📌 0
TL;DR Forecasting oncogene amps & tumour suppressor dels is feasible! This can refine risk stratification and anticipate treatment resistance, paving the way for earlier, smarter and more personalised cancer care. There is much more in the preprint so check it out! 13/
10.06.2025 14:15 — 👍 0 🔁 0 💬 1 📌 0

MET amps cause EGFRi resistance in ~25% of NSCLCs. Forecasting MET amp in 33 EGFR-mutant NSCLC tumours treated with osimertinib showed high-risk patients had shorter PFS & OS. This can be used to flag candidates for upfront EGFR+MET inhibition (eg MARIPOSA trial) 12/
10.06.2025 14:15 — 👍 0 🔁 0 💬 1 📌 0

Currently, LGGs are classified into 4 WHO risk groups. CDK4/PDGFRA amps and CDKN2A dels are linked with poor prognosis but under utilised. Forecasting these facilitates a risk upgrade of 9% of IDHmut-non-codel cases while maintaining median survival times across WHO groups 11/
10.06.2025 14:15 — 👍 0 🔁 0 💬 1 📌 0
Encouraging right? We then applied our approach to two clinical scenarios where forecasting specific genetic changes might unlock new clinical opportunities: risk stratification of low-grade glioma (LGG) and anticipation of osimertinib resistance in lung cancer 10/
10.06.2025 14:15 — 👍 0 🔁 0 💬 1 📌 0

Next we tested longitudinal pairs forecasting at the early time point (before driver amp) and testing at the latter. In prostate, we predicted AR amp (linked to ADT resistance) in pretreatment samples. In NSCLC, we predicted HIST1H3B amp (exclusive to metastases) in primaries 9/
10.06.2025 14:15 — 👍 0 🔁 0 💬 1 📌 0
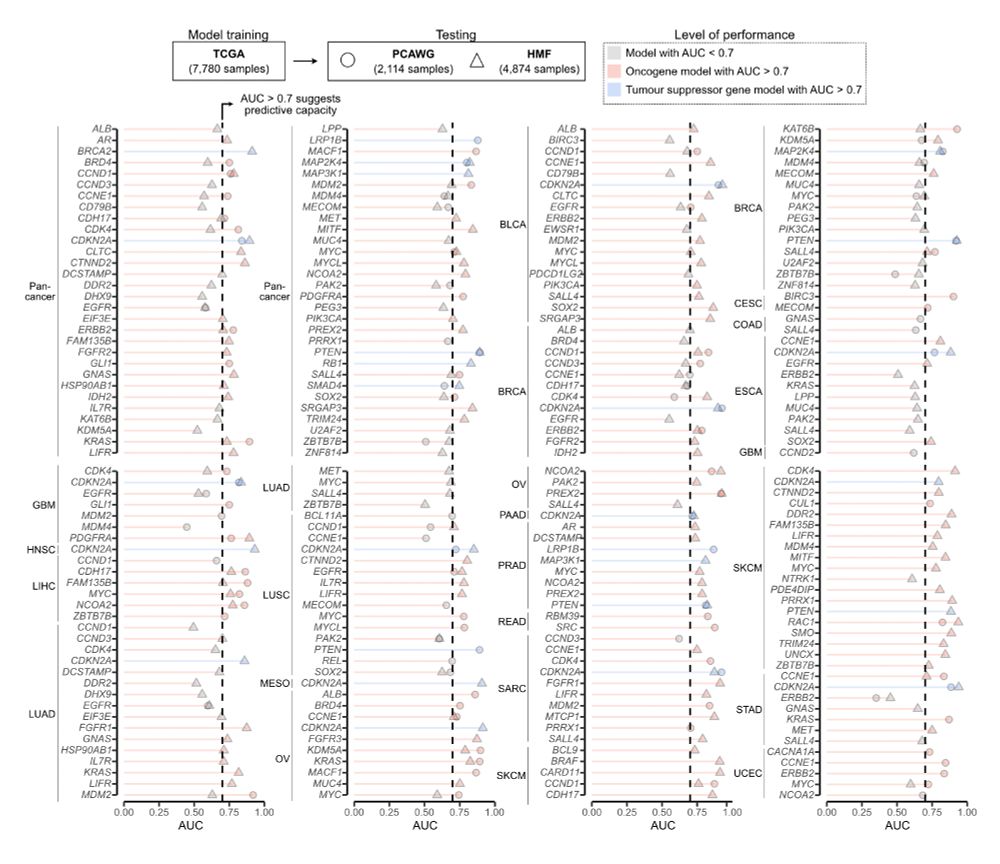
First, we tested performance on two independent cohorts: PCAWG: 2,114 primaries; HMF: 4,784 metastases. 147 of the 241 models showed AUC > 0.7 across both datasets 8/
10.06.2025 14:15 — 👍 0 🔁 0 💬 1 📌 0
We trained this model for 241 drivers using 7,880 TCGA samples across 33 tumour types. But do these models work? 7/
10.06.2025 14:15 — 👍 0 🔁 0 💬 1 📌 0

Challenge 3: forecasting in a clinical setting. Solution: binarise predictions and only use standard genomic test data as input. We designed guidelines to apply and (if needed) train the model + optimize thresholds for binary risk classification (high vs low) 6/
10.06.2025 14:15 — 👍 0 🔁 0 💬 1 📌 0

Challenge 2: growth rates of mutant vs non-mutant cells (and thus selection) cannot be easily determined in a clinical context. Solution: approximate selection coeffs using driver amp/del frequency at a population-level (supported by recent work showing s≈fβ) 5/
10.06.2025 14:15 — 👍 0 🔁 0 💬 1 📌 0

Challenge 1: amp/del rates cannot be readily measured from an input genome. Solution: approximate using a steady-state probability of locus-specific copy number change over tumour lifetime. We adapted our previous CIN signatures (CX bit.ly/42SVA3i) for this 4/
10.06.2025 14:15 — 👍 0 🔁 0 💬 1 📌 0
Simple model? Seemingly so. But estimating mutation rate and selection coefficients from tumour DNA alone, especially for DNA copy number, is tough! We faced a number of challenges: 3/
10.06.2025 14:15 — 👍 0 🔁 0 💬 1 📌 0
Chancellor's Fellow (~Asst. Prof.) University of Edinburgh
| Math modelling for somatic genomics | https://michaeldnicholson.github.io
whole genome molecular oncology, genome graphs and graph genomes, cancer karyotype assembly, cancer chromosome folding, breakage fusion bridge cycles, tyfonas, and other HSRs
Assistant Professor at WashU, St. Louis. Interested in damage and breaks...
https://www.vermalab.org/
LMCB Group Leader at UCL. Cis-regulatory control of cell fate choice in development.
https://delaslab.com/
Here for #bioinformatics / #computationalBiology of #Genomics, #Oncology and #Virology. Also loves #dataViz and open-source software.
Immunoinformatics at Mcgranahan lab @uclcancer Ex @eapm_bsc @bsc_cns @carrerasIJC - Stirring the pan at @eltallaret @embassat - he/him
We study DNA topology, the action of topoisomerses and DNA break repair to understand genome organization, dynamics and stability @cniostopcancer.bsky.social
https://repairome.bioinfo.cnio.es
Burkard Lab | Holden Comprehensive Cancer Center | University of Iowa
Studying chromosomal instability, mitotic regulation & cancer evolution
Directed by Dr. Mark E. Burkard
Group leader @gustaveroussy.fr. Computational Oncology 💻 | Cancer genomics 🧬 | Clonal Hematopoiesis and Leukemia🩸
Assistant Editor for Clinical Cancer Research published by AACR | PhD in Genetics and Cancer Biology | Views are my own | Header by Nathan Pyle
Tea addict and scientist, in that order. Incoming Group Leader, Dept of Oncology, University of Oxford. All views my own.
Marie Skłodowska-Curie fellow at ISEM Montpellier. Physicist studying complex biological systems - ecosystems, cancer, immune networks and microbial communities.
Studying cellular #stress responses, particularly #senescence and its impact on #immune response, #ageing and #cancer, #tumorigenesis at Cancer Research UK CI @cruk-ci.bsky.social, University of Cambridge @cam.ac.uk
Website: naritalab.com
Professor of Cancer Evolution. UCL Cancer Institute. Interested in cancer genomics, bioinformatics, and somatic evolution.
https://www.ucl.ac.uk/medical-sciences/divisions/cancer/our-research/cancer-genome-evolution
Postdoc @uwgenome 🐶
PhD Genetics @uarizona 🐱
BA Chemistry @brynmawrcollege 🦉
Population Genomics, Machine Learning, Cancer Evolution 🧬💻
📍Seattle, WA & Vancouver, BC
https://www.linkedin.com/in/linhnhtran/
Biochemist 🧪| Empowering Life Scientists and Science Communicators to Achieve Their Career Goals | UCLA PhD + NIH Alum | Pharma R&D @GSK + @AstraZeneca 💊 | #SciComm Director ✍️| Career Coach | Star Trek | Sherlock Holmes | 🎹 | https://larrymillerphd.com
Professor of Molecular Genetics at the University of Oxford. Gastroenterology doctor.
Richard Blackwell Pharsalia Professor of Colorectal Surgery | Group Leader, NDS (ORCRB) | Fellow of Linacre | University of Oxford
Professor of Neuropathology, Director Brain Tumour Research Centre, Blizard Institute, Queen Mary University of London
#brain #tumours #epigenetics #stemcells #models …ah yes and #hiking enthusiast #dog-lover













