From systems operators to systems architects
Going up a level from data generation to think about the data systems we design and embed
Announcing the Diffuse Project! We're unlocking protein dynamics through diffuse X-ray scattering - the overlooked signal that could revolutionize how we understand protein motion. seemay.substack.com/p/from-syste...
12.08.2025 16:21 — 👍 9 🔁 6 💬 1 📌 1

The results of the ISCB Leadership vote are in!
Terms for the ISCB President-Elect and ISCB Vice President will start on January 21, 2026.
13.08.2025 13:08 — 👍 10 🔁 2 💬 1 📌 1
I just got the notice that all the FlyBase people at Harvard, including me, will be laid off on October 12. I'm devastated.
11.08.2025 18:12 — 👍 376 🔁 225 💬 66 📌 50

Scaling down protein language modeling with MSA Pairformer
Recent efforts in protein language modeling have focused on scaling single-sequence models and their training data, requiring vast compute resources that limit accessibility. Although models that use ...
Excited to share work with
Zhidian Zhang, @milot.bsky.social, @martinsteinegger.bsky.social, and @sokrypton.org
biorxiv.org/content/10.1...
TLDR: We introduce MSA Pairformer, a 111M parameter protein language model that challenges the scaling paradigm in self-supervised protein language modeling🧵
05.08.2025 06:29 — 👍 91 🔁 41 💬 1 📌 1
Stay tuned for details on the 6th edition of MLSB, officially happening this December in downtown San Diego, CA!
28.07.2025 15:41 — 👍 14 🔁 7 💬 0 📌 0
That's a wrap! The results of the first #cryoEM heterogeneity challenge are up on biorxiv!
biorxiv.org/content/10.110
23.07.2025 21:43 — 👍 45 🔁 21 💬 3 📌 4


Everyone working in a STEM field should read this - ‘writing is thinking’
www.nature.com/articles/s44...
23.07.2025 18:07 — 👍 123 🔁 39 💬 4 📌 5

New Study Reveals Subclasses of Autism by Linking Traits to Genetics
New Study Reveals Subclasses of Autism by Linking Traits to Genetics on Simons Foundation
#FlatironCCB scientists Natalie Sauerwald, Olga Troyanskaya and colleagues leveraged @sparkforautism.bsky.social data to reveal four distinct groups that link autism-related traits with underlying genetics. Read more: www.simonsfoundation.org/2025/07/09/n... #science #biology #autismresearch
21.07.2025 14:22 — 👍 2 🔁 2 💬 0 📌 0
We hope this update will significantly expand the types of researchers + compute systems that can use D-SCRIPT! Manuscript with benchmarks + implementation details coming hopefully soon, and thanks again so much to Daniel for his hard work pushing this out!
22.07.2025 18:39 — 👍 0 🔁 0 💬 0 📌 0
Along with a bunch of other quality-of-life updates (including a mode specifically designed for host x pathogen/symbiont interactions! 🦠), v0.3.0 will power the sorts of comparative genomics and network analyses that we imagined when we first designed D-SCRIPT.
22.07.2025 18:39 — 👍 1 🔁 0 💬 1 📌 0
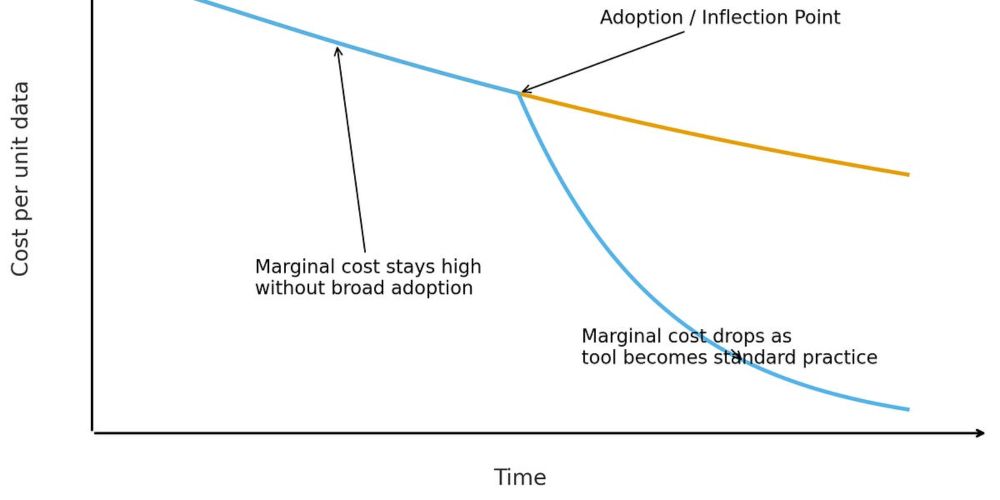
BMPI makes D-SCRIPT fully parallel in both embedding loading and inference, and can distribute inference to an arbitrary number of available GPUs. At the same time, the blocked inference procedure means that memory usage can be decreased arbitrarily low (with a small trade-off in speed).
22.07.2025 18:39 — 👍 0 🔁 0 💬 1 📌 0
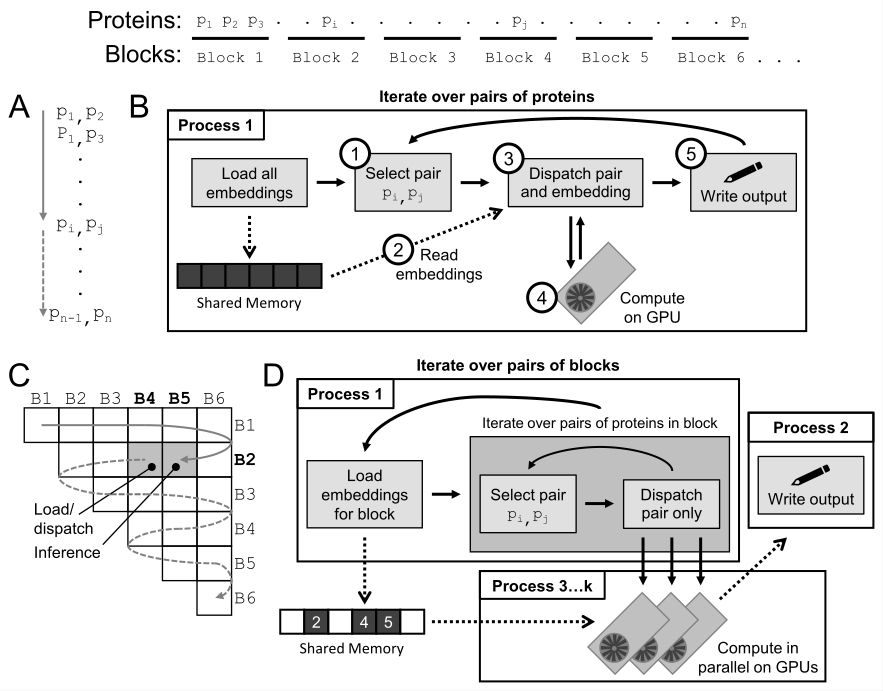
In v0.3.0, we solve *both* of these issues, enabling efficient inference on both personal computers and multi-GPU HPC systems. The secret? Our new Blocked Multi-GPU Parallel Inference (BMPI) procedure, led by Daniel Schaffer (github.com/schafferde).
22.07.2025 18:39 — 👍 0 🔁 0 💬 1 📌 0
This ends up being extremely wasteful, since D-SCRIPT still made predictions in serial on a single GPU; inference was not possible on personal systems, but much of the compute capacity of these large machines was sitting unused. Whole-genome all-by-all prediction could still take upwards of a week!
22.07.2025 18:39 — 👍 0 🔁 0 💬 1 📌 0
Despite this, one of the most common user issues we heard about was that pre-loading embeddings achieved speed by trading off massive memory usage, restricting D-SCRIPT inference at whole-genome scale only to large HPC-like machines.
22.07.2025 18:39 — 👍 0 🔁 0 💬 1 📌 0
We designed D-SCRIPT to be extremely high-throughput, and the most common use case we've seen is whole-proteome all-by-all prediction (this use case is at the core of our recent 🎼philharmonic work!)
22.07.2025 18:39 — 👍 0 🔁 0 💬 1 📌 0
The short film "Molecular Duet" brings the dynamic world of microtubules to life. A collaboration between @flatironinstitute.org researcher Mahsa Mofidi and filmmaker Anne Sofie Nørskov. Watch in full: www.simonsfoundation.org/2025/07/16/w... #science #biology #film #FlatironCCB
17.07.2025 20:13 — 👍 1 🔁 2 💬 0 📌 0
From #FlatironCCB scientist Bargeen Turzo and filmmaker Grace Zhang, "How to Connect Two Bodies" is an experimental documentary exploring the invisible choreography in meaningful connections of objects across various scales. Watch in full: www.simonsfoundation.org/2025/07/09/w... #science #film
16.07.2025 16:14 — 👍 1 🔁 1 💬 0 📌 0
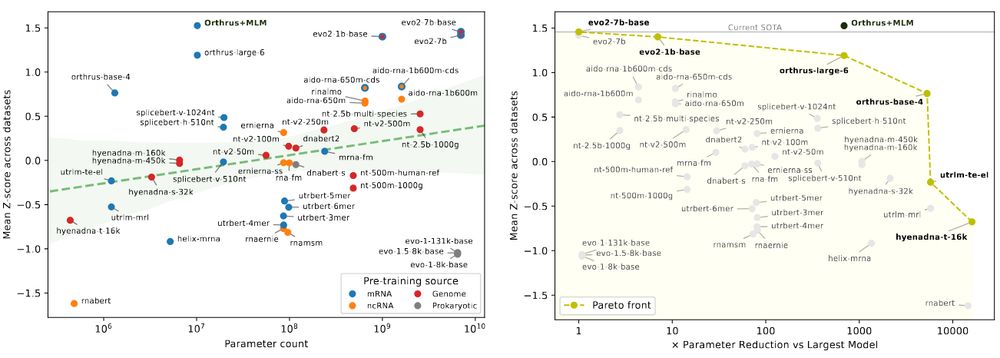
We're excited to release 𝐦𝐑𝐍𝐀𝐁𝐞𝐧𝐜𝐡, a new benchmark suite for mRNA biology containing 10 diverse datasets with 59 prediction tasks, evaluating 18 foundation model families.
Paper: biorxiv.org/content/10.1...
GitHub: github.com/morrislab/mR...
Blog: blank.bio/post/mrnabench
15.07.2025 18:41 — 👍 21 🔁 5 💬 1 📌 0
Today in the journal Science: BioEmu from Microsoft Research AI for Science. This generative deep learning method emulates protein equilibrium ensembles – key for understanding protein function at scale. www.science.org/doi/10.1126/...
10.07.2025 18:10 — 👍 106 🔁 49 💬 1 📌 3
Wow, congratulations!🎉
09.07.2025 03:10 — 👍 1 🔁 0 💬 1 📌 0
CASP is the main reason the protein structure prediction technology and research field advanced over the last 30 years. And the main reason AI based methods have been accepted and widely applied in biology. So shortsighted of NIH to postpone or even halt funding. John Moult is a scientific hero.
02.07.2025 22:36 — 👍 41 🔁 14 💬 1 📌 0
One thing that really bothers me with the new "virtual cell" terminology is that it is currently largely focused on a very narrow definition of models that can predict effects of trans perturbations (gene dosage, drugs etc) on gene expression. 1/
28.06.2025 10:38 — 👍 103 🔁 30 💬 1 📌 0
Extremely cool work here!
25.06.2025 19:26 — 👍 1 🔁 0 💬 0 📌 0
You can start using RocketSHP 🚀 now on GitHub, and we're actively working on better models, improved analysis tools, and ways to link prediction/regression-type models with sampling-based approaches. And if you have any large-scale complex trajectories you want to share, my DMs are always open! 😁
23.06.2025 20:42 — 👍 1 🔁 0 💬 0 📌 0
Infrastructure for learning and representing protein motions from structural data
Computer scientist at I2SysBio-CSIC. Transcriptomics, long-reads, multiomics, tool developer. Promoter of Green Science. Finally happy!
AI-powered platform for proteomics and beyond. Fast, intuitive analysis for scientists. Built by top-tier engineers and scientists. Check us out at www.tesorai.com.
Assistant Professor at University of Illinois at Chicago (UIC) in Biological Sciences focused on macromolecular assemblies.
https://research.nvidia.com/labs/genair/
Principal Research Scientist at NVIDIA | Former Physicist | Deep Generative Learning | https://karstenkreis.github.io/
Opinions are my own.
My lab at Stanford studies human population genetics and complex traits.
I write Buzzer, a college basketball newsletter, at eamonnbrennan.com. I do other stuff, too.
An independent college basketball outlet.
basketunderreview.com
A global community of researchers dedicated to the understanding of proteins.
PhD student @sangerinstitute
Solve Biology
Head of Generative Genomics, Wellcome Sanger Institute, Cambridge, UK
Systems + Synthetic Biology, CRG, Barcelona
http://barcelonacollaboratorium.com http://allox.bio https://www.sanger.ac.uk/programme/generative-and-synthetic-genomics/
Research scientist at @GoogleDeepMind passionate about AI, genomics and biology.
PhDing at the Sanger Institute, i'm evolving every day
Post-doc @UniofOxford w/ @mmbronstein.bsky.social. Into Geometry ∩ Generative Models. @mila-quebec.bsky.social Affiliate member. Phd from @mila-quebec.bsky.social / McGill.
website: https://joeybose.github.io/
A new scientific institution for curiosity-driven biomedical science and technology.