
Undergraduates
Rosetta Commons Research Experience for Undergraduates (REU) AI for Biomolecular Structure Prediction and Design Interns in this geographically-distributed REU program participate in research using…
Final stretch to apply for undergraduate summer internships in the Rosetta Commons! Come design proteins, develop AI and physics-based methods to model biomolecules, and impact health, materials, and sustainability! Application deadline is Sunday Feb 1.
rosettacommons.org/education/reu/
30.01.2026 16:09 — 👍 3 🔁 4 💬 0 📌 0
New lab preprint - Common and rare variant studies for the same trait identify different genes and here Diederik Laman Trip developed a protein network AI enconding to investigate if traits studied by different approaches converge on the same molecular pathways
www.biorxiv.org/content/10.6...
29.01.2026 08:12 — 👍 19 🔁 10 💬 2 📌 0
YouTube video by Alice Ting
Alice Ting Rosettacon keynote 2025
Video introduction to our new “Conformational Biasing” method for computational design of mutations that bias proteins towards desired conformational states
CB part starts at 14:55
Thanks to Peter Cavanagh and Andrew Xue – amazing graduate students who co-led this work
08.01.2026 19:15 — 👍 24 🔁 8 💬 1 📌 1
Congrats!
08.01.2026 03:11 — 👍 1 🔁 0 💬 0 📌 0
Modern biology research is biased towards investigating genes that are widely conserved and present in humans. What about genes that ARE widely conserved but NOT present in humans? Can genes missing from humans tell us something about what makes our biology different from that of other animals? 1/8
31.12.2025 19:29 — 👍 85 🔁 28 💬 10 📌 1
Know when to co-fold'em
This is the official web page for the James Fraser Lab at UCSF.
I'm really excited to break up the holiday relaxation time with a new preprint that benchmarks AlphaFold3 (AF3)/“co-folding” methods with 2 new stringent performance tests.
Thread below - but first some links:
A longer take:
fraserlab.com/2025/12/29/k...
Preprint:
www.biorxiv.org/content/10.6...
29.12.2025 22:25 — 👍 72 🔁 30 💬 5 📌 2
RFpeptides made it to the cover of Nature Chemical Biology December issue!
Credit to Stephen Rettie for leading the work and making this very cool graphic!
03.12.2025 22:58 — 👍 5 🔁 1 💬 0 📌 0
Very happy to share our latest led by the relentless @shawnfayer.bsky.social! We developed multiplexed assays to measure variant effects across diverse genetic and cell contexts in stem cell and differentiated cells. 🧵 👇
22.11.2025 17:08 — 👍 4 🔁 1 💬 0 📌 0


New preprint! We measured temperature- and pH-induced aggregation for over 18,000 natural and de novo designed protein domains!
19.11.2025 21:16 — 👍 121 🔁 42 💬 4 📌 3


Congratulations to the newly minted Dr. Brittney Voigt! After defending a fantastic thesis spanning work in my lab and the @edwardmarcotte.bsky.social lab.
19.11.2025 02:03 — 👍 21 🔁 3 💬 2 📌 1
Last week, the research of Rui Gao and lab was published by
@Science.org and featured on the issue's cover! Dr. Rui Gao is an Assistant Professor in Biological Sciences and Chemistry.
science.org/doi/10.1126/...
23.10.2025 16:14 — 👍 1 🔁 2 💬 0 📌 0

The Gao lab at #UIC reports a breakthrough in volumetric fluorescence imaging, featured on the cover of Science Magazine!
Their new technique, VIPS (Volumetric Imaging via Photochemical Sectioning), enables reconstruction of 3D images of whole tissues with unprecedented detail.
16.10.2025 22:26 — 👍 9 🔁 4 💬 1 📌 0

Beyond Wolbachia—Can a small molecule control insect reproduction?
Kaur et al. demonstrate reduced histone acetylation as a key mechanism underpinning
Wolbachia’s paternal-effect embryonic lethality trait in Drosophila melanogaster.
Recapitulation of this trait by in...
#Wolbachia has puzzled scientists with its power to rewire insect reproduction. What if I tell you that we found one of the keys Wolbachia use to rewire its host AND a small molecule inhibitor uses this key to mimic what this microbe has mastered for millions of years.
www.cell.com/cell-reports...
03.10.2025 12:57 — 👍 63 🔁 26 💬 2 📌 3
Congrats Qian www.science.org/doi/full/10....
29.09.2025 13:19 — 👍 1 🔁 1 💬 0 📌 0
I am excited to share our new preprint on the CAGE complex, a mysterious hollow protein complex that I first saw years ago while surveying Tetrahymena ciliary lysate www.biorxiv.org/content/10.1... #cilia #protistsonsky 🧬🧪
23.09.2025 17:08 — 👍 167 🔁 54 💬 7 📌 13

10 Rules for a Structural Bioinformatic Analysis
The Protein Data Bank (PDB) is one of the richest open‑source repositories in biology, housing over 277,000 macromolecular structural models alongside much of the experimental data that underpins thes...
Structural bioinformatics is incredibly powerful on its own or when paired with theory or experiment. One of the PDB's superpowers isn’t from one structure, but comparing many to uncover folds, binding sites, and subtle conformational shifts. chemrxiv.org/engage/chemr...
11.09.2025 14:28 — 👍 54 🔁 15 💬 1 📌 2
Excited for you to join us!
10.09.2025 01:37 — 👍 2 🔁 1 💬 1 📌 0

🚨The Bits to Binders Competition has concluded!🧬
One year ago we gathered scientists from around the world to design and submit protein binders that cause immune cells to target and eliminate CD20+ tumors
Spoiler: They work!
02.09.2025 14:45 — 👍 5 🔁 2 💬 1 📌 1
Exciting to see our protein binder design pipeline BindCraft published in its final form in @Nature ! This has been an amazing collaborative effort with Lennart, Christian, @sokrypton.org, Bruno and many other amazing lab members and collaborators.
www.nature.com/articles/s41...
27.08.2025 16:14 — 👍 305 🔁 109 💬 14 📌 11

Scaling down protein language modeling with MSA Pairformer
Recent efforts in protein language modeling have focused on scaling single-sequence models and their training data, requiring vast compute resources that limit accessibility. Although models that use ...
Excited to share work with
Zhidian Zhang, @milot.bsky.social, @martinsteinegger.bsky.social, and @sokrypton.org
biorxiv.org/content/10.1...
TLDR: We introduce MSA Pairformer, a 111M parameter protein language model that challenges the scaling paradigm in self-supervised protein language modeling🧵
05.08.2025 06:29 — 👍 97 🔁 43 💬 1 📌 1
Also, I would like to thank the NSF and NIH for funding, and DOE Argonne, NSF ACCESS, and Indiana JetStream2 for computational resources.
29.07.2025 20:58 — 👍 1 🔁 0 💬 0 📌 0
There are lots of other results in the manuscript so please check it out. I definitely want to thank the team of Miles Woodcock-Girard, Erin Claussen, and Samantha Fischer who did the work.
29.07.2025 20:58 — 👍 1 🔁 0 💬 1 📌 0

OFD1 disease mutations (pink) cluster at the OFD1(green)-FOPNL(orange) interface.
Finally, we utilize our network to build structural models of a ciliary protein complex linked to oral-facial-digital syndrome. We identified several human pathogenic mutations at the interface of OFD1 and FOPNL (CEP20), pointing to a mechanism of pathology.
29.07.2025 20:58 — 👍 2 🔁 0 💬 2 📌 0

The fraction of AlphaFold3 protein interaction models with high-confidence pDockQ pairs.
Encouraged by this, we prioritized ~2,500 high-confidence DirectContacts2 protein pairs and ran them through AF3. We see excellent enrichment of high quality models consistent with the AF2 models.
29.07.2025 20:58 — 👍 2 🔁 0 💬 1 📌 0
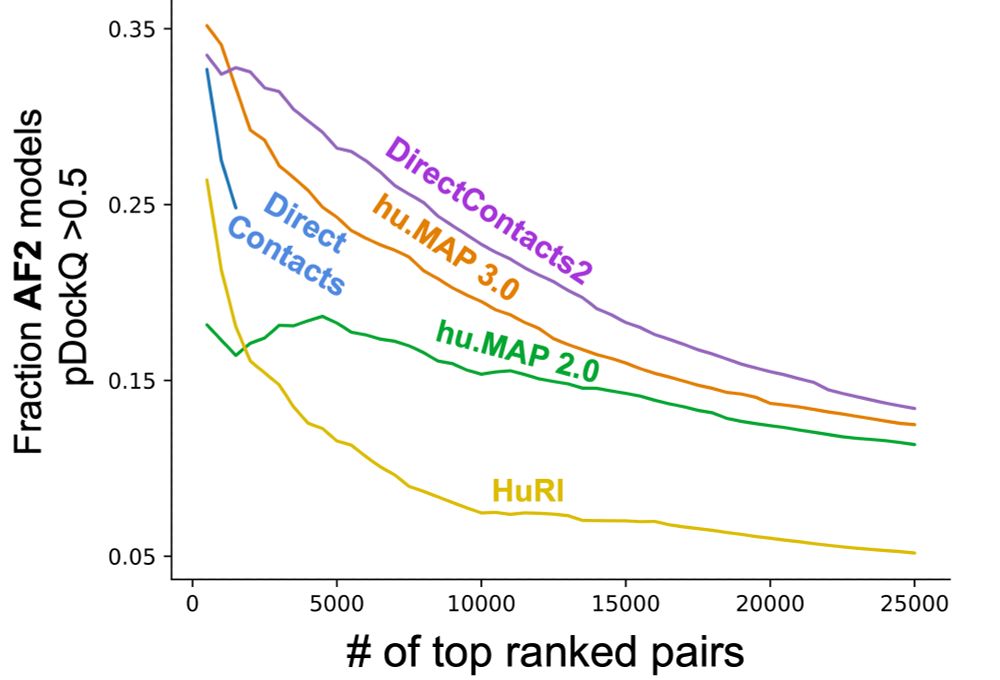
Comparison of enrichment for high-confidence pair interfaces out of the set of top confident pairs for DirectContacts2 and other networks. For each set, the fraction of high-confidence interfaces (pDockQ ≥ 0.5) out of their top predictions was calculated for increasing numbers of top predictions.
First, using a compendium of computed structural models for evaluation, we see our DirectContacts2 network outperforms other networks at prioritizing protein pairs for AlphaFold2 modeling.
29.07.2025 20:58 — 👍 3 🔁 1 💬 1 📌 0
Previous work by @bioinfo.se and @pedrobeltrao.bsky.social, among others, prioritized pairs using protein interaction networks, including using our hu.MAP complex map.
29.07.2025 20:58 — 👍 1 🔁 0 💬 1 📌 0
Assistant professor in the biology department at the University of Illinois Chicago. Lab website https://walkerlab-uic.github.io/. All opinions, spelling mistakes, and grammatical errors are still my own.
Neurobiologist and Assistant Professor at the University of Illinois Chicago studying sensory cells and neural circuits. #Neuroscience and #Olfaction.
https://www.zak-lab.org
The Theoretical Biology Reading Group meets monthly at the University of Illinois Chicago to discuss texts pertaining to fundamental issues in the life sciences. theoreticalbiology.github.io
Scientist interested in structural biology, vaccine design, antibodies, protein engineering… pretty much everything
Systems Bio, comparing things to each other, protein language models, plants, Asst prof UArizona MCB
RCSB PDB (RCSB.org) promotes a structural view of biology. Funded by NSF, NCI, NIAID, NIGMS, NIH, and DOE
Biologist especially interested in cilia, signaling and development!
Chemist (Food, 3D printing), Professor (American University), Author (Chemistry in Your Kitchen)
Structural biologist studying cilia and ciliopathies.
Full stack developer who occasionally wears a lab coat. Currently a machine learning researcher at Manifold Bio.
Londoner. CBRN expert, particularly nerve agents. Freeman of the City of London. Liveryman. "Legal Juggernaut". Anglican. Ex Verger. Author. Fellow of the Royal Historical Society. Scourge of llamas. Former US Army. Former USSS. #NAFO Fella
Discover the Languages of Biology
Build computational models to (help) solve biology? Join us! https://www.deboramarkslab.com
DM or mail me!
Solve difficult biological problems using ML/AI.
Cancer Risk Assessment - “Catch Cancer Early”.
Perturbation Biology - computational cell biology - design new combo therapies.
Protein design - function and structure.
curious about all things bio & solving tricky structural puzzles / mlbio for protein design, engineering & drug disco / cytokine immunologist / stanfordberkelyucsf & dnaxgenentech / recovering twit
Our laboratory studies the extracellular matrix #ECM in health and disease using #Proteomics and #CompBiology | Founder of the Matrisome Project @matrisome.bsky.social | https://nabalab.uic.edu
Computational Structural Biologist
Assistant Professor @VanderbiltMPB
wankowiczlab.com
(she/her)
Past: UCSF, Dana-Farber, Broad Institute, UMass Amherst
#NewPI @UIC
Studying Host-Symbiont-Virus Interactions in 🪰 and 🦟
#Wolbachia #Spermatogenesis #Embryogenesis #ChromatinBiology #Epigenetics #Microscopy
She/her
Ramón y Cajal Junior Group Leader @BSC_CNS 🇦🇷🇪🇸🏳️🌈 ENFJ | Bioinformatics | 3DSIG Co-Chair | @fi_uner, @exactas_uba, @mpi_bpc, @embl, @dkfz alumnus | @iscb BoD | Opinions my own
protein geek, founder and CEO of AI Proteins @aiproteins.com
https://linkedin.com/in/cdbahl



















