If you like transposons...
If you you love genome editing...
Or if you just like random bird animations,
we have the paper for you!
We (@kedmonds.bsky.social et al) are happy to share our work turning a songbird retrotransposon into a genome editing tool. 🐣 (1/n)
03.07.2025 03:20 — 👍 45 🔁 16 💬 4 📌 2
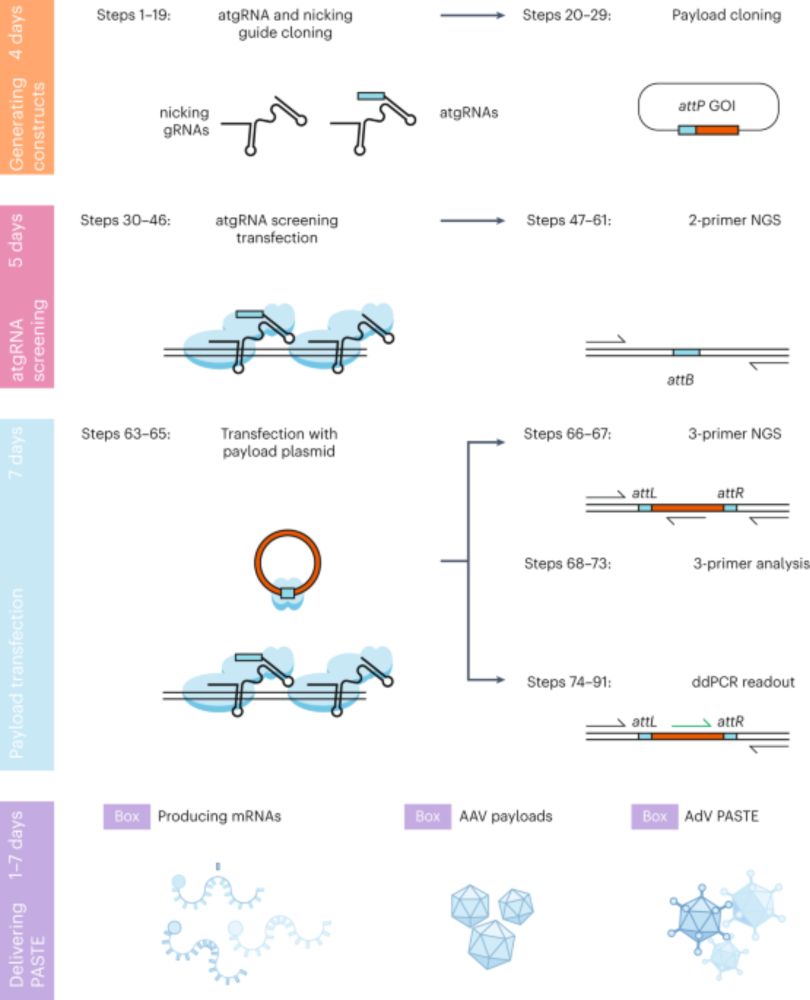
STITCHR, a tool that can make virtually any kind of edit in mammalian genomes including the scarless multi-kilobase scale insertions. Plasmids here www.addgene.org/browse/artic...
10.04.2025 20:06 — 👍 10 🔁 3 💬 0 📌 0
We're looking into it :) the differences in UTR dependencies between in vitro and in cells is interesting. Big fan of your work!
10.04.2025 13:13 — 👍 0 🔁 0 💬 0 📌 0
Lastly, I’m very thankful of my excellent co-first authors Lukas Villiger, Justin Lim, Masa Hiraizumi, and mentors Omar and Jonathan. We are so grateful for our fantastic ongoing collaborations with Hiroshi Nishimasu. Thanks to all co-authors for their invaluable contributions!
09.04.2025 17:15 — 👍 2 🔁 0 💬 1 📌 0
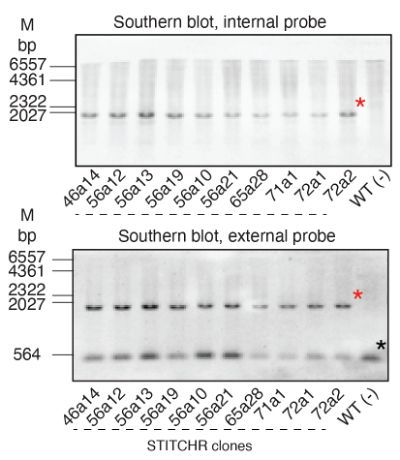
This was a really fun project which I’m excited is finally out. There were some unique challenges on the way, including learning how to Southern blot to address a reviewer’s concern. The blot got buried in the supplemental, but it took me months, so here it is in all its glory
09.04.2025 17:14 — 👍 2 🔁 0 💬 1 📌 0
There’s much more data in the paper, so please check it out! We also showed insertion by delivering RNA templates, insertion in different cell types, in vitro characterization of STITCHR, mapping of nicking sites, PacBio sequencing of long insertions, offtarget mapping and more
09.04.2025 17:14 — 👍 2 🔁 0 💬 1 📌 0
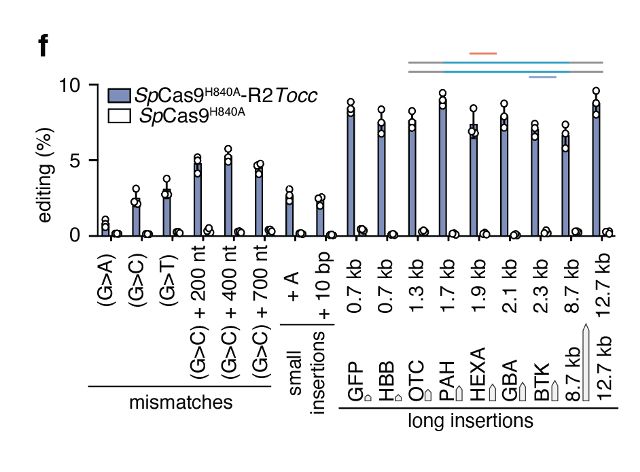
STITCHR can insert whatever is between its homology arms. We can efficiently install a diverse range of edits, including substitutions, deletions and large insertions of at least 12.7kb with high fidelity
09.04.2025 17:14 — 👍 1 🔁 0 💬 1 📌 0
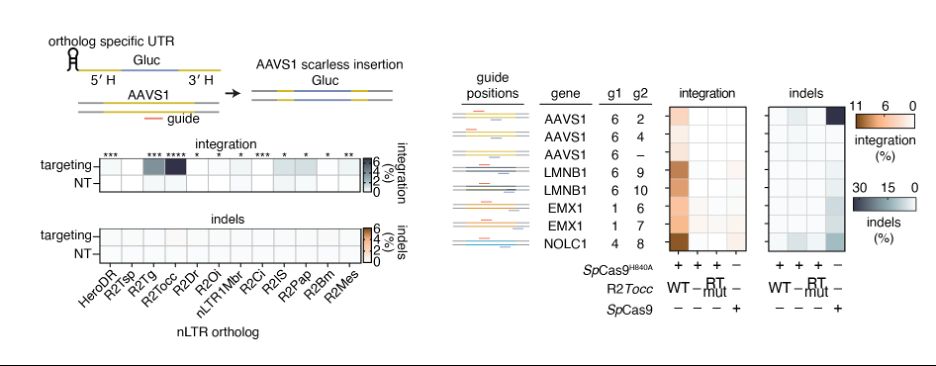
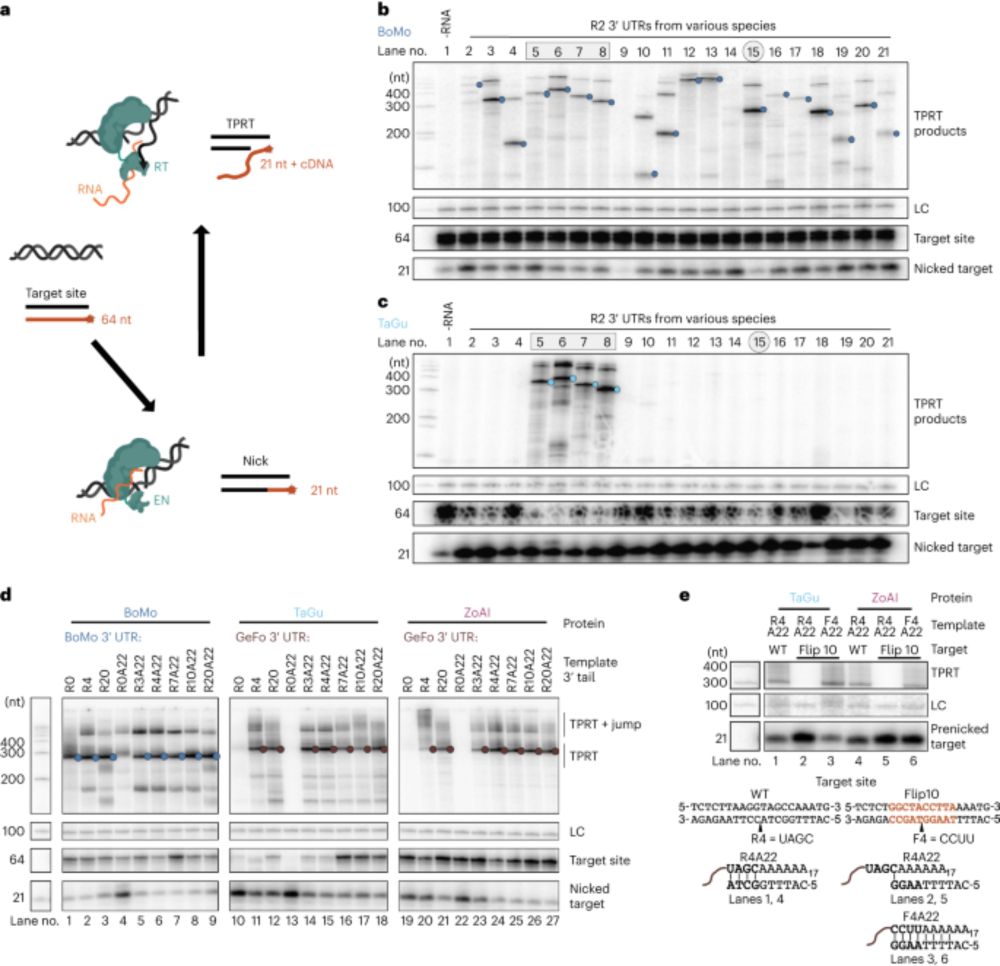
We tested a panel of R2s for their ability to perform scarless insertion at AAVS1, identifying R2Tocc as our best orthologue due to high on-target insertion and low 28S insertion. After screening sgRNA panels and homology arms, we could insert efficiently at multiple genomic loci
09.04.2025 17:14 — 👍 0 🔁 0 💬 1 📌 0
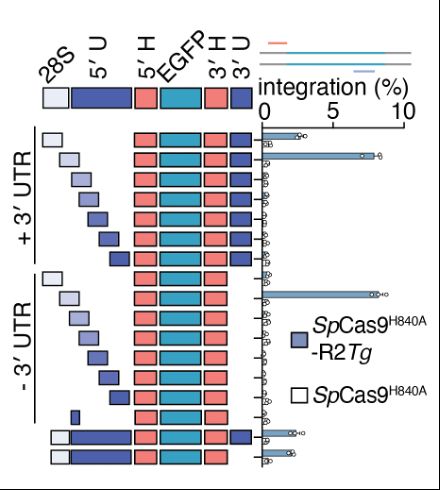
We found the architecture of the RNA cargo to be critical. The 3’ UTR is dispensable in cells, however, the homology arms and a small section of the 5’ UTR are essential, including a short sequence homologous to its natural 28S insertion site.
09.04.2025 17:13 — 👍 0 🔁 0 💬 1 📌 0
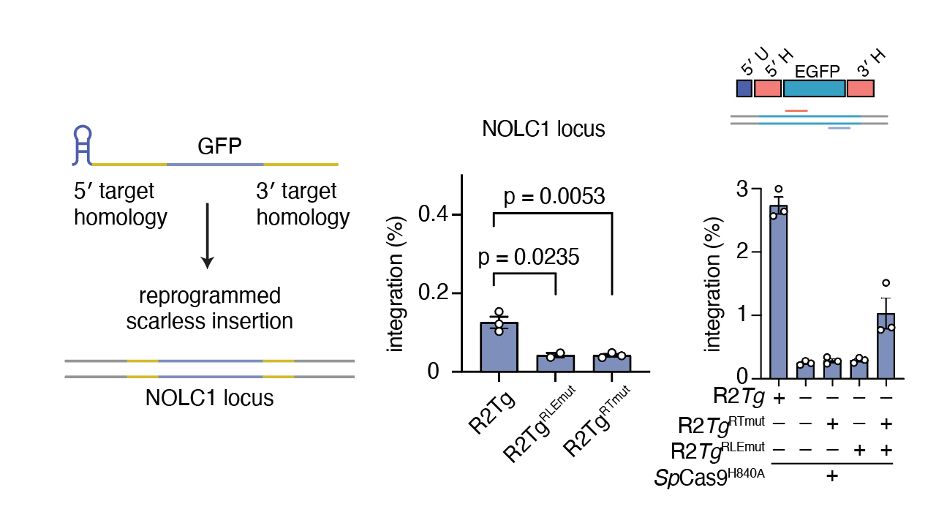
Swapping out homology arms for NOLC1 sequences led to the natural reprogramming of R2Tg, inserting a GFP at NOLC1, which was improved with nCas9-assited retargeting. We called this system Site-specific Target-primed Insertion via Targeted CRISPR Homing of Retroelements (STITCHR).
09.04.2025 17:13 — 👍 1 🔁 0 💬 1 📌 0
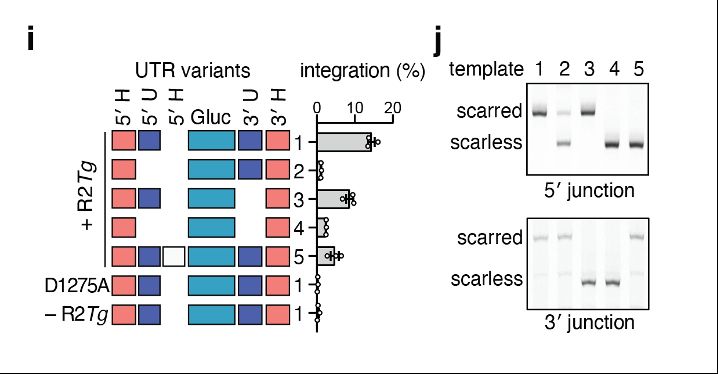
To understand cargo constraints, we permuted R2Tg’s RNA template, deleting and/or re-ordering the UTR and homology arm architecture. Interestingly, deletion of the UTRs internal to the homology arms were still functional and resulted in scarless cargo insertion!
09.04.2025 17:13 — 👍 0 🔁 0 💬 1 📌 0
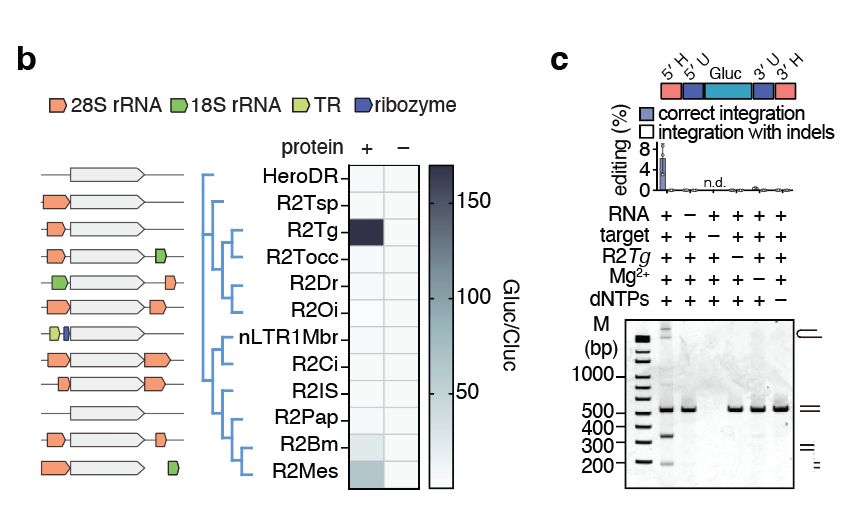
We selected a diverse set of R2s and tested if they could insert non-natural cargoes at their natural sites in human cells. We identified an R2 from the zebra finch Taeniopygia guttata (R2Tg) with high insertion activity in cells and in vitro.
09.04.2025 17:13 — 👍 0 🔁 0 💬 1 📌 0
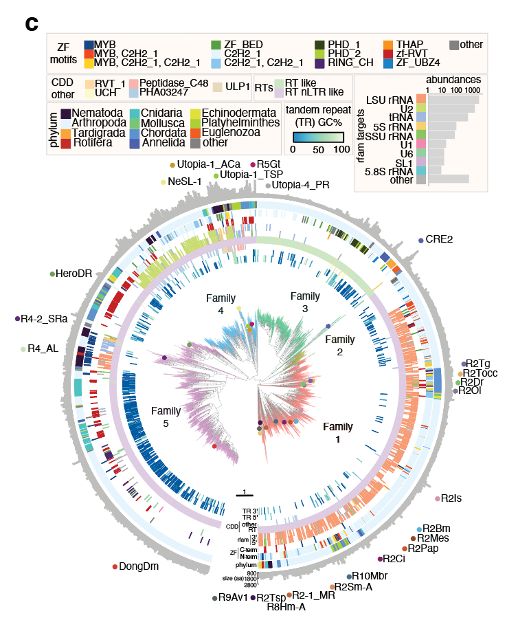
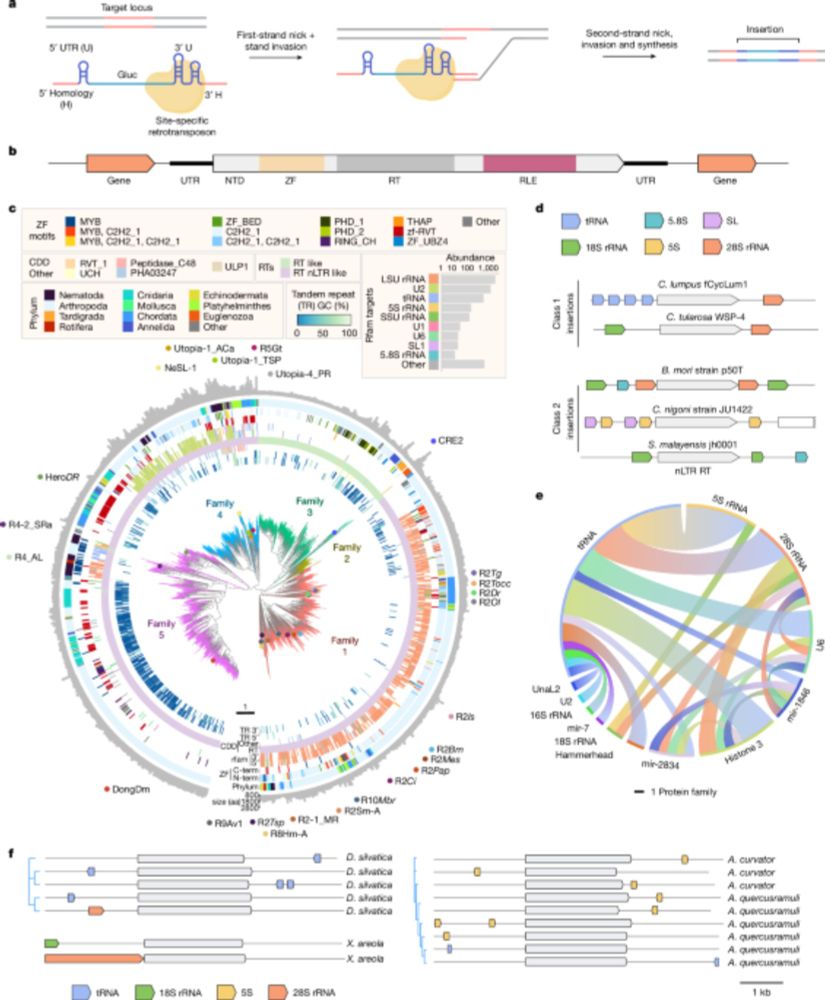
We were interested in whether R2s could be reprogrammed in cells. We searched the natural world, uncovering 8,248 orthologues and their site preferences. Excitingly, we discovered instances of R2s acquiring novel insertion sites during evolution, a natural form of reprogramming!
09.04.2025 17:12 — 👍 0 🔁 0 💬 1 📌 0
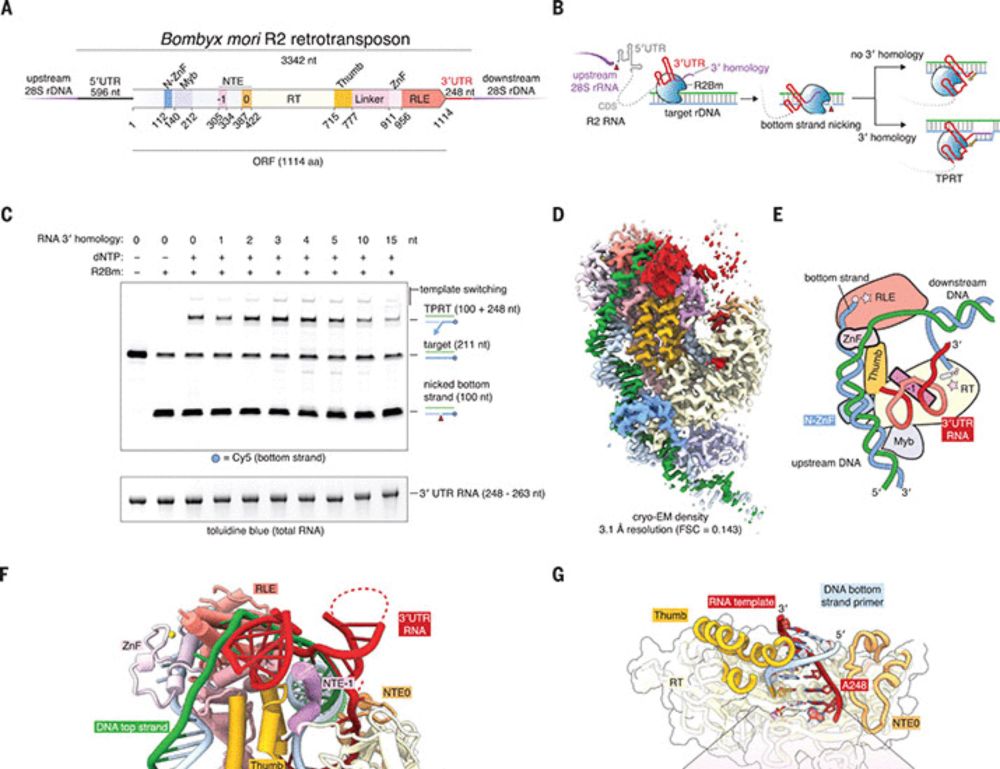
In the AbuGoot lab, we were intrigued by the R2 retrotransposon class of mobile genetic elements. Foundational work by Thomas Eikbush and colleagues showed a Target-Primed Reverse Transcription mechanism of insertion, priming with RNA homology arms.
09.04.2025 17:11 — 👍 0 🔁 0 💬 1 📌 0
Postdoc in the Zhang lab @ Broad Institute. Formerly in the Elowitz lab @ Caltech.
Née Ke-Huan Chow.
I ❤️ SynBio and cats.
Assistant Professor, Columbia University | Mobile elements, Structural biology and Genome engineering | http://thawanilab.org
AITHYRA is a new dynamic research institute for biomedical AI in Vienna. AITHYRA seeks to build Europe’s premier institute for AI-driven biological and medical research, uniting computer scientists, engineers, and biologists in a collaborative environment.
Senior PhD student 🥼 Villunger lab at CeMM 🇦🇹 Epitranscriptomics | Molecular medicine | Immunology | Manžel 🇨🇿 'Fins up 🐬🏈
keen on reverse transcription | also keen on spliceosomes | cryoEM dabbler
Assistant Member @MSKCC
Formerly post-doc @MIT
Formerly-formerly PhD @MRC_LMB
Formerly formerly-formerly Otago Uni
Kiwi 🥝 🇳🇿
https://wilkinsonlab.bio/
International, independent, interdisciplinary research institute. Integrates basic research and clinical expertise for innovative diagnostics and therapeutics.
🔗 http://www.cemm.oeaw.ac.at/
Cell metabolism & mitochondria | HFSP-alumni & Postdoc at MSKCC
Assistant Professor @ Princeton MolBio |
From transposons to telomeres and back | bicycles and biochemistry
https://ghanim.lab.princeton.edu
Located at the University of California-Berkeley at the Innovative Genomics Institute.
Purveyors of Microbial Ecology, Bioinformatics, & Nanogeoscience.
Reposts or likes≠endorsements.
https://www.banfieldlab.com/
Bioengineer PhD student @ Harvard
Duke ‘24
Proud rodeo mom, soccer mom, baseball mom, hockey mom, constitutional conservative.
A project of The Center For Scientific Integrity https://centerforscientificintegrity.org/
Daily newsletter http://eepurl.com/bNRlUn
Database http://retractiondatabase.org
Tips team@retractionwatch.com (better than @ replies)
News, commentary and research coverage from the international monthly journal publishing the highest quality of work in all areas of neuroscience.
https://www.nature.com/neuro/
Immunologist | HFSP Postdoctoral Fellow @RiceLaboratory | PhD @bergthalerlab | opinions are my own.
Professor, UW Biology / Santa Fe Institute
I study how information flows in biology, science, and society.
Book: *Calling Bullshit*, http://tinyurl.com/fdcuvd7b
LLM course: https://thebullshitmachines.com
Corvids: https://tinyurl.com/mr2n5ymk
he/him
In-depth, independent reporting to better understand the world, now on Bluesky. News tips? Share them here: http://nyti.ms/2FVHq9v
Functional genomics @ Broad Institute. Screen all the things!
Professor and Chair, Dept Microbiology Cornell Ithaca. Interested in genome evolution and mobile DNA, especially Tn7 and CRISPR-Cas transposition systems