Our “Less is more” story is out! 🚀 We reveal how gene loss shaped the adaptive evolution of pandemic Vibrio parahaemolyticus, highlight a potentially general mechanism across bacteria. Huge thanks @danielfalush.bsky.social @campy-bara.bsky.social @jaimemurtaza.bsky.social & all coauthors
27.08.2025 13:06 — 👍 10 🔁 5 💬 0 📌 0
What's left to discover: What are the viscous liquids in which Molassodon hunts, kills and devours bacterial prey? We now have many more tools to help us find out, and to understand the trade offs for different bacterial strategies. All comments welcome, thank you for reading.
30.06.2025 04:23 — 👍 0 🔁 0 💬 0 📌 0
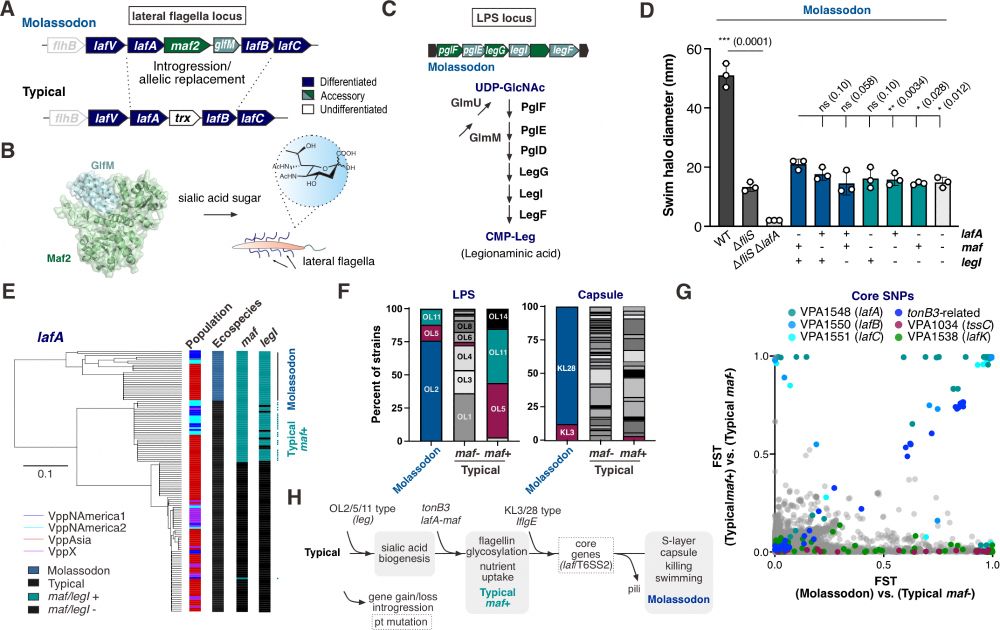
We also identify a convergent intermediate form and are also able to provide details about the molecular and evolutionary assembly of the Molassodon genotype, revealing a key role for lateral flagella glycosylation.
30.06.2025 04:23 — 👍 1 🔁 0 💬 1 📌 0
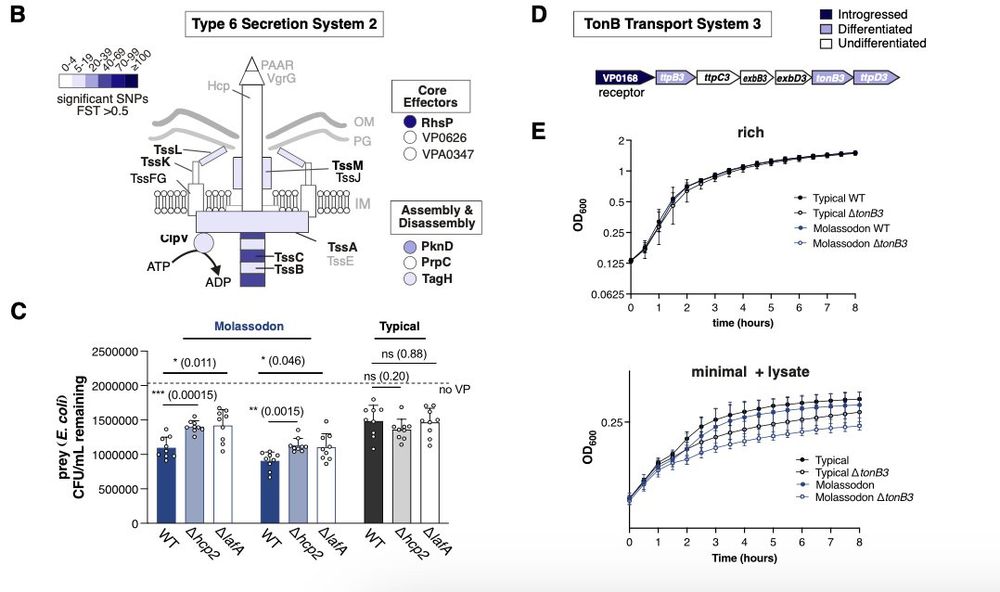
Upregulated genes provide clues about the ecological advantage of fast swimming in viscous liquids. A Type 6 secretion system and nutrient uptake genes are upregulated, allowing the bacteria to kill and devour bacterial prey.
30.06.2025 04:23 — 👍 0 🔁 0 💬 1 📌 0
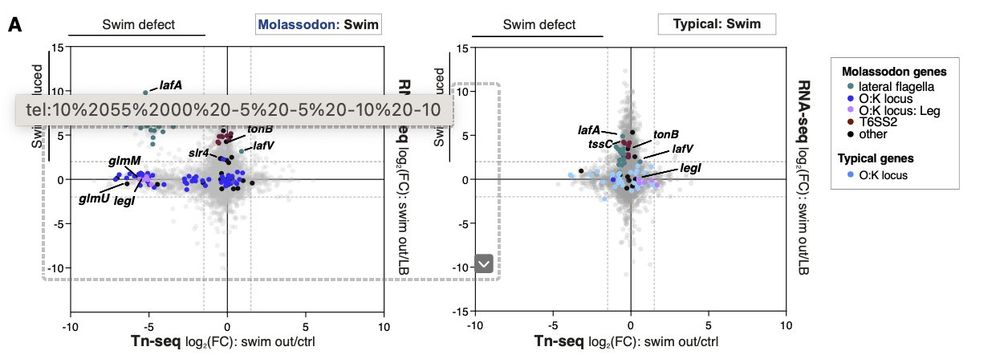
We use TNseq and RNA seq to show that 2/3 of ecospecies loci are either essential for this enhanced swimming or are unregulated along with it.
30.06.2025 04:23 — 👍 1 🔁 0 💬 1 📌 0
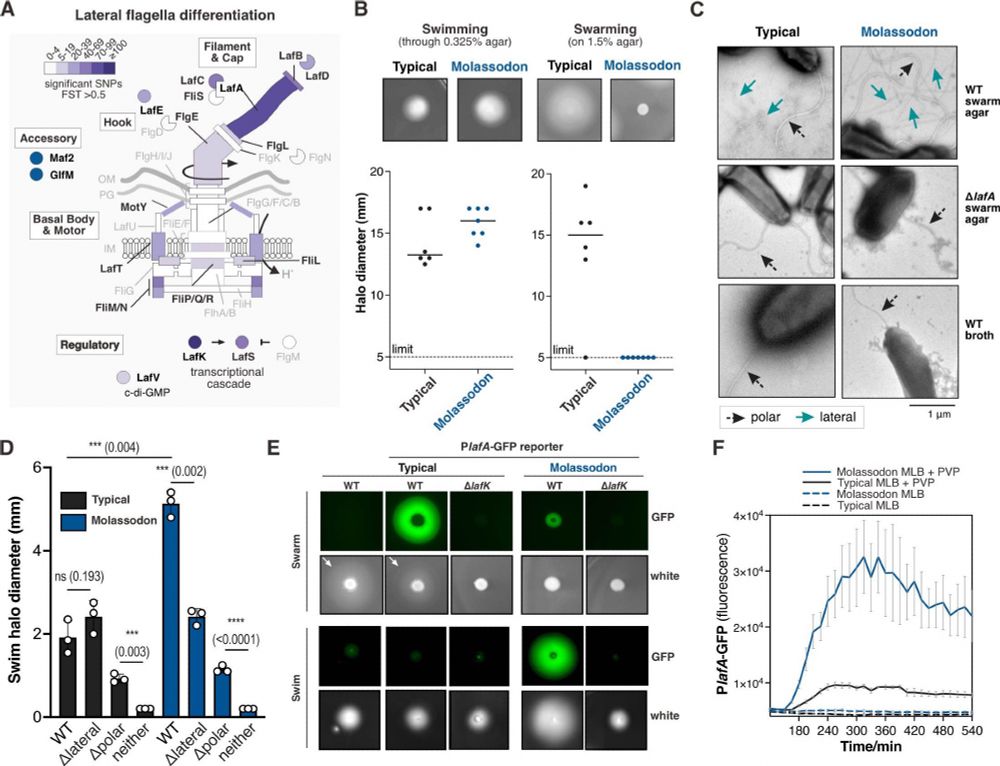
Bacterial ecospecies are powerful tools for molecular biological, evolutionary and ecological investigation because all of the differentiated genes are working together. Molassodon have abandoned swarming on surfaces and instead use their lateral flagella to swim faster in viscous liquids.
30.06.2025 04:23 — 👍 0 🔁 0 💬 1 📌 0
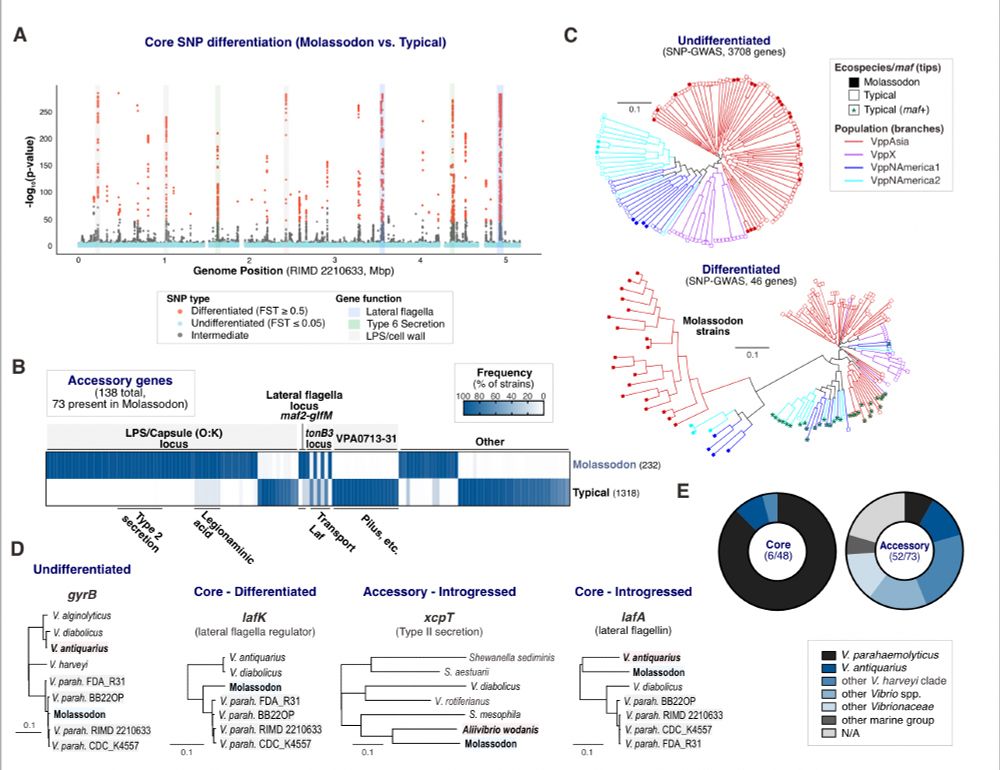
Bacterial ecospecies are complex adaptations on an otherwise freely recombining background. Molassodon has evolved via introgression of accessory genes and a few core genes from other Vibrio species, as well as point mutation.
30.06.2025 04:23 — 👍 0 🔁 0 💬 1 📌 0

Molassodon! Excited to post my research group's first laboratory project. We identify a phenotype and a putative ecological strategy for the Molassodon ecospecies of Vibrio parahamolyticus, thanks to the hard work of our lab team led by @campy-bara.bsky.social www.biorxiv.org/content/10.1...
30.06.2025 04:23 — 👍 8 🔁 6 💬 1 📌 1
There is every chance to make a recovery from these cuts and to leave science in a healthier place than it was, for example if the opposition to them galvanizes a common sense of purpose across American society - don't lose sight of that!
18.06.2025 12:37 — 👍 1 🔁 0 💬 1 📌 0
We have lived within a period of unprecedented stability and.. should keep some sense of perspective about this. Second World War and cultural revolution caused incomparably greater upheaval, for example, silly to compare, but many things were better than ever a couple of human generations later.
18.06.2025 12:37 — 👍 0 🔁 0 💬 1 📌 0
I agree it's terrible, my sympathies, but I do think that hyperbolae, "multiple generations" are not helpful. In my parents lifetime, I can think of much worse things that have happened and been recovered from.
17.06.2025 06:02 — 👍 1 🔁 0 💬 1 📌 0
follow me Graham!
24.03.2025 08:30 — 👍 0 🔁 0 💬 0 📌 0
Thank you for reading our thread, please read the preprint too! your comments on all aspects are extremely welcome.
28.02.2025 06:05 — 👍 0 🔁 0 💬 0 📌 0
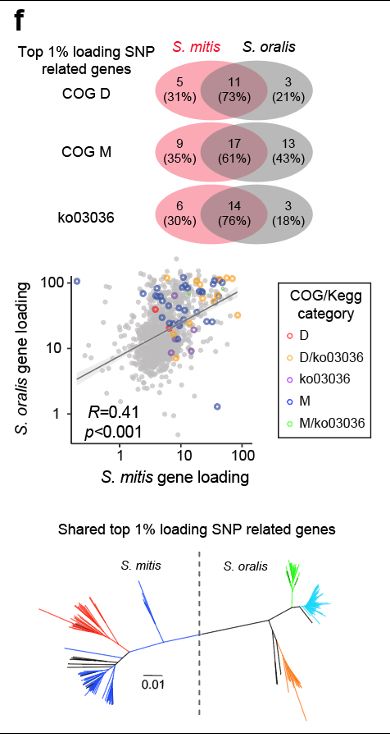
Fascinatingly, their convergence in ecospecies differentiation between the two streptococcus species, involving the same genes and gene categories (cell division related), despite independent origins. Different genera find different but predictable paths for adaptive differentiation.
28.02.2025 06:05 — 👍 0 🔁 0 💬 1 📌 0
The first two bacterial ecospecies we discovered, in Heliocobacter pylori and vibrio parahaemolyticus are characterized by high differentiation restricted to a small fraction of the genome. The streptococcal ones have progressed further, consistent with the older age of the species.
28.02.2025 06:05 — 👍 0 🔁 0 💬 1 📌 0
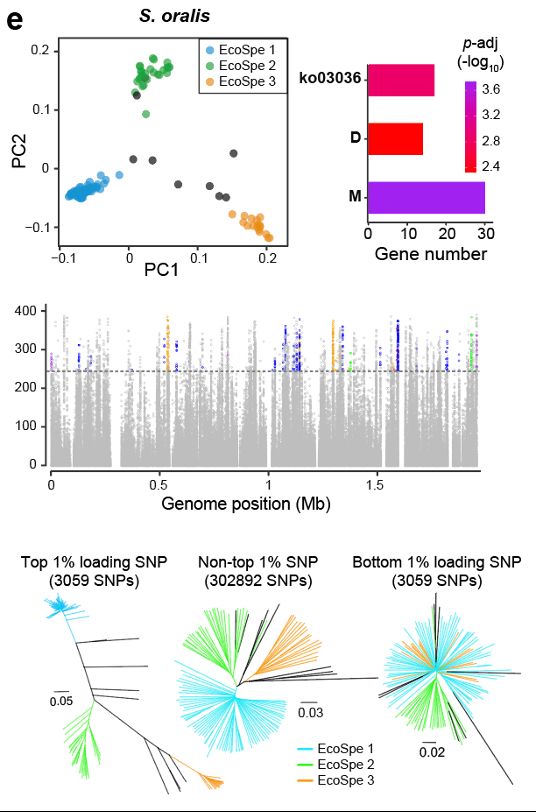
And the first bacterial ecospecies threesome, in Streptococcus oralis.
28.02.2025 06:05 — 👍 0 🔁 0 💬 1 📌 0
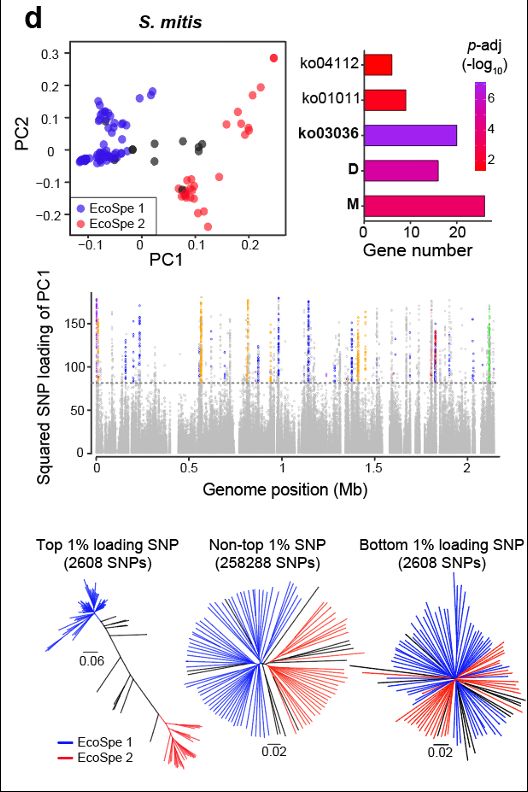
We illustrate this concretely by discovering a third bacterial ecospecies pair in Streptococcus mitis.
28.02.2025 06:05 — 👍 0 🔁 0 💬 1 📌 0
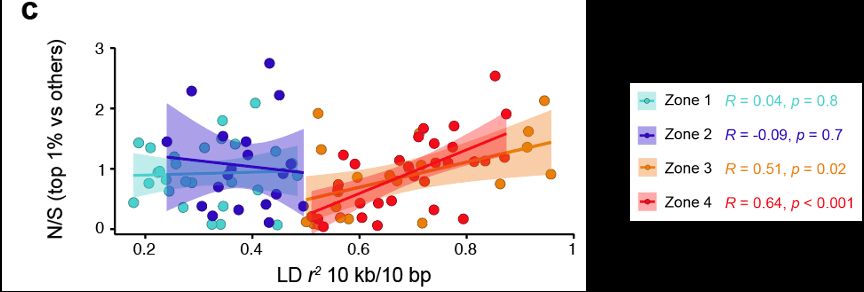
Why? Well what happens is that for species which maintain large population sizes for long time periods, natural selection becomes increasingly important in structuring diversity and this generates long range LD.
28.02.2025 06:05 — 👍 2 🔁 0 💬 1 📌 0
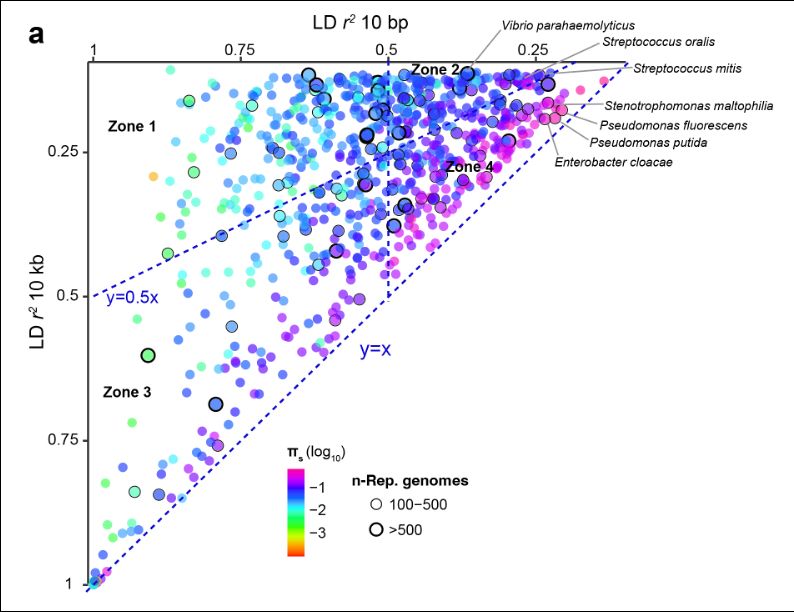
Last and not least, we provide an answer to the question of why panmictic bacterial species are rare. A well-mixed bacterial species should have low LD at all genetic distances, but there are no such species. Instead we find that the most diverse species have intermediate long-range LD.
28.02.2025 06:05 — 👍 0 🔁 0 💬 1 📌 0
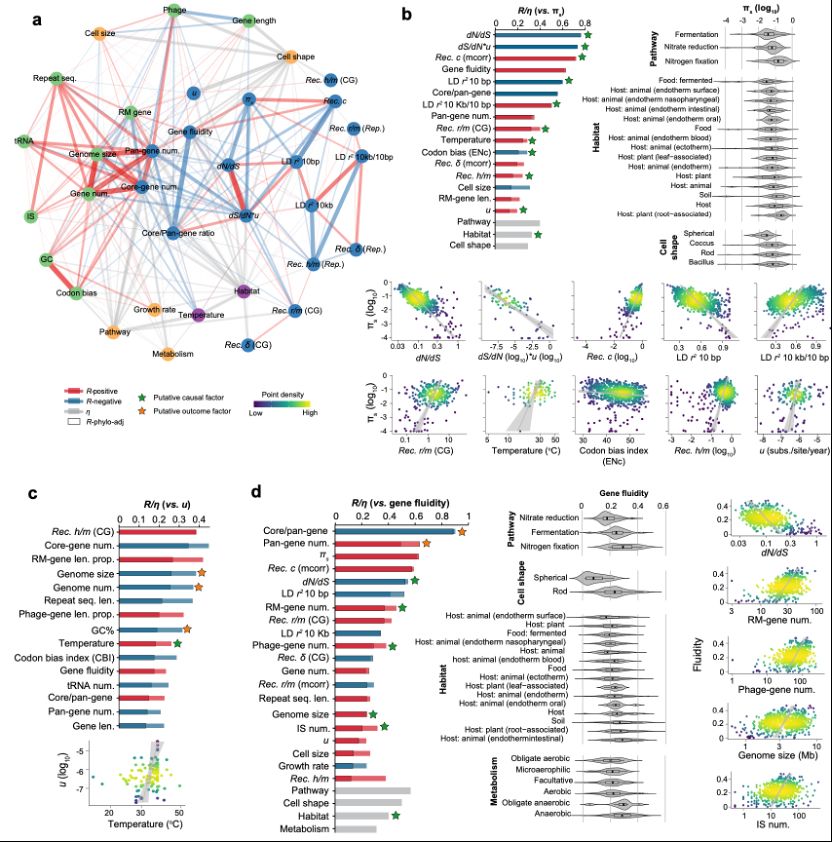
And there are curious findings too, like the fact that the genome fluidity of rod shaped bacteria is more than double that of spherical ones. What can you see in the data? Or what would you like to see? Let us know!
28.02.2025 06:05 — 👍 0 🔁 0 💬 1 📌 0
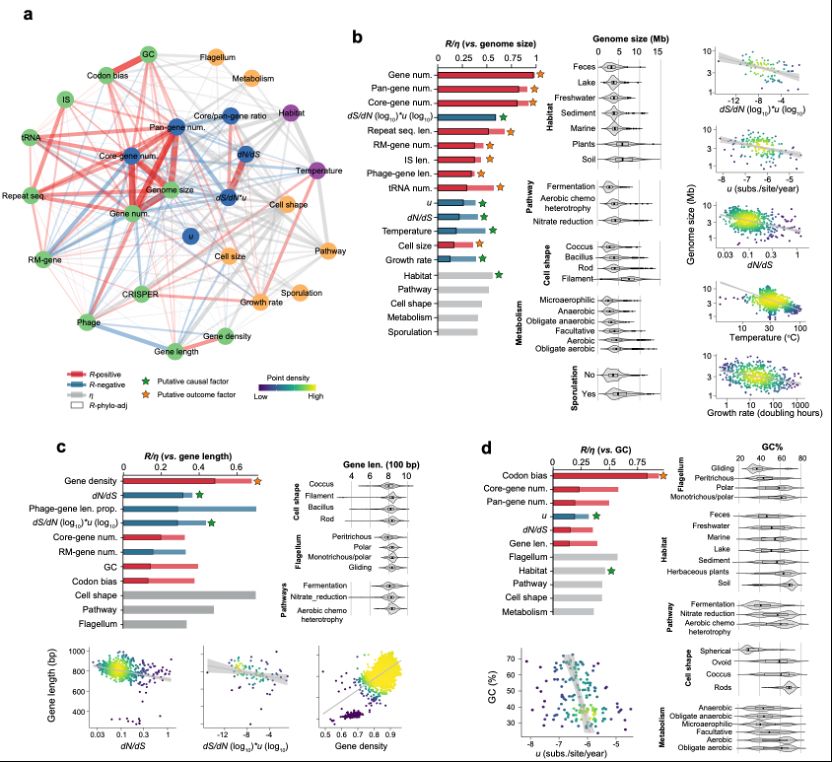
We are able to use the population genetic data to address classic questions such as what determines the number of genes? It seems to be a balance between deletion bias and selection favoring increasing genome complexity.
28.02.2025 06:05 — 👍 1 🔁 0 💬 1 📌 0
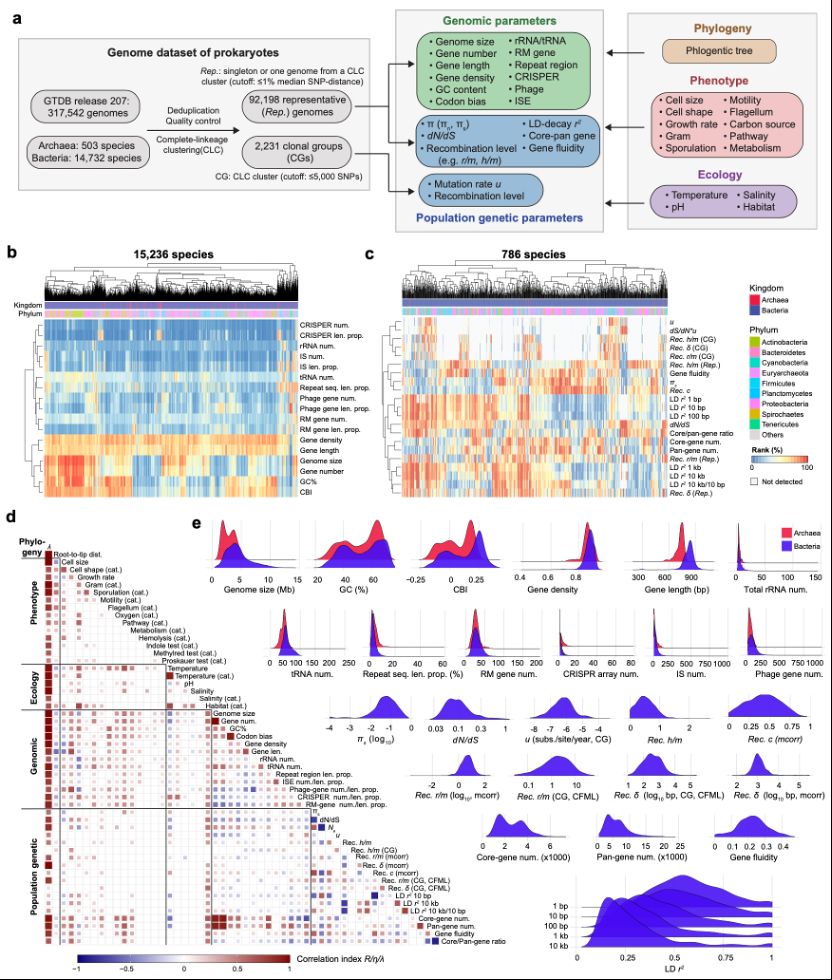
Why has it taken us so long to update it? Like fine wine, it has matured. Unlike fine wine, it has also grown in scale. We now analyze population genetic data from nearly 800 species, instead of twenty something and genomic data from 15000. Figure 1 presents an overview of genomic parameters.
28.02.2025 06:05 — 👍 0 🔁 0 💬 1 📌 0

Macrogenetic atlas of prokaryotes
Macrogenetics investigates the patterns and predictors of intraspecific genetic variation across diverse taxa, offering a framework to address longstanding evolutionary hypotheses. Here, we present a ...
Very interesting new preprint from the Falush lab in Shanghai, describing an effort to parameterize a large number of microbial and population (phylo)genetic traits for >15k species, then examining their interactions.
Loads to dig up & possibly quite impactful!
🔗 www.biorxiv.org/content/10.1...
27.02.2025 05:45 — 👍 6 🔁 3 💬 0 📌 0
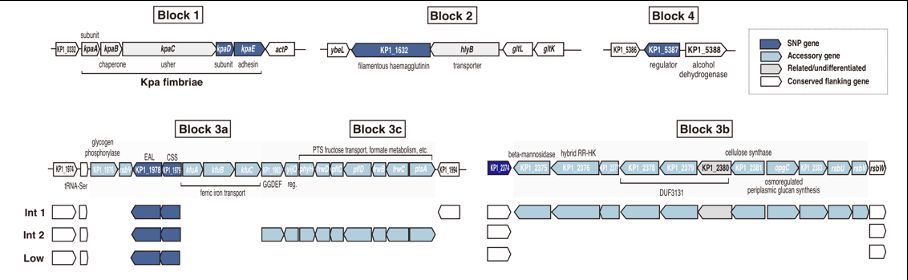
So the million dollar question, what is the trait? Is there a trait? We don't know. The top loading genes provide some hints (number 1 is a fimbria locus). Really interested to hear your thoughts and suggestions, thanks for reading and reposting :).
27.01.2025 08:34 — 👍 0 🔁 0 💬 0 📌 0
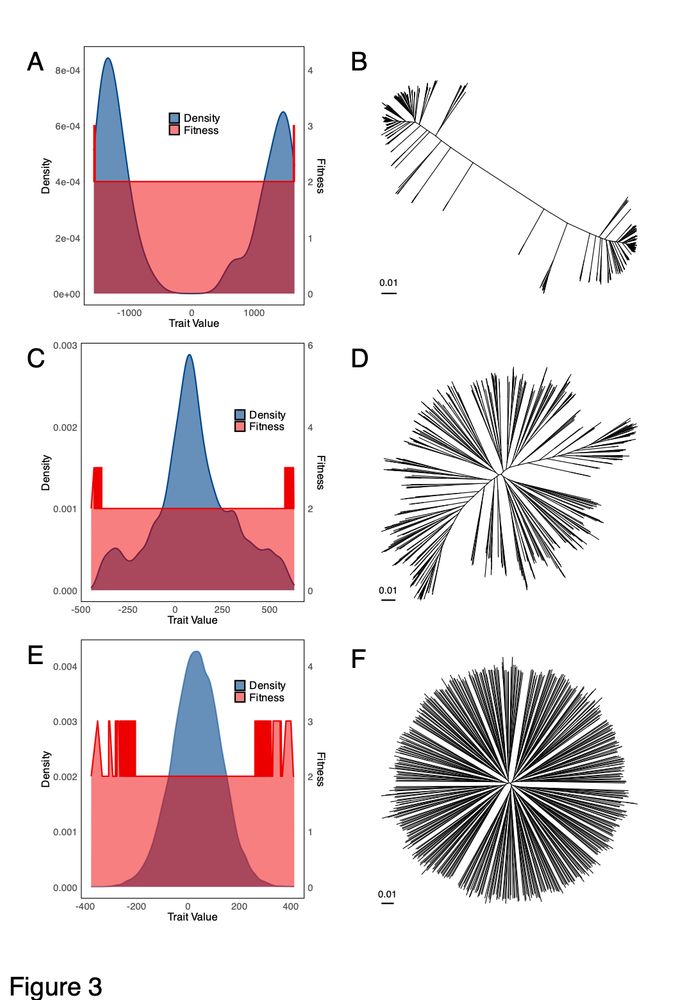
We propose instead that the structure is due to diversifying selection on a quantitative trait. The trait is influenced by many genes, and the most extreme genotypes have a small advantage. For low recombination rates, the species splits into two. For intermediate rates get an ecocline.
27.01.2025 08:34 — 👍 0 🔁 0 💬 1 📌 0
So what biological factors might explain this component of variation? Klebsiella recombines frequently, meaning that clonal relationships cannot obviously account for this species-wide structure. Population structure might, except you find the same lineages everywhere.
27.01.2025 08:34 — 👍 0 🔁 0 💬 1 📌 0
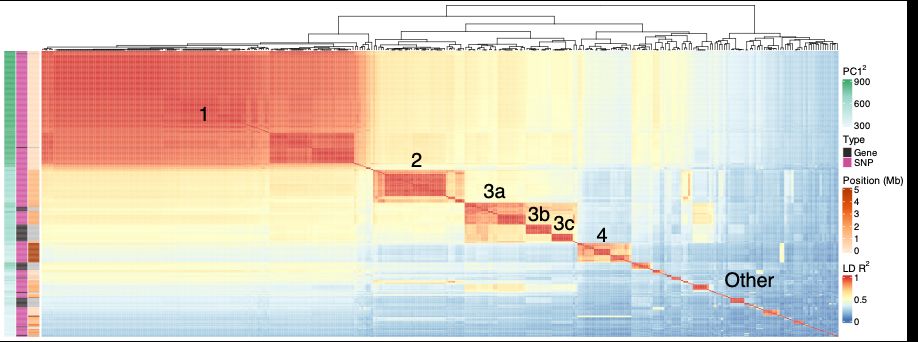
Another way to visualize this is by looking at pairwise Linkage Disequilibrium (LD) between the most differentiated genes. This shows that PC1 captures the structure of association with PC1 values explaining pairwise LD between genes well.
27.01.2025 08:34 — 👍 1 🔁 0 💬 1 📌 0
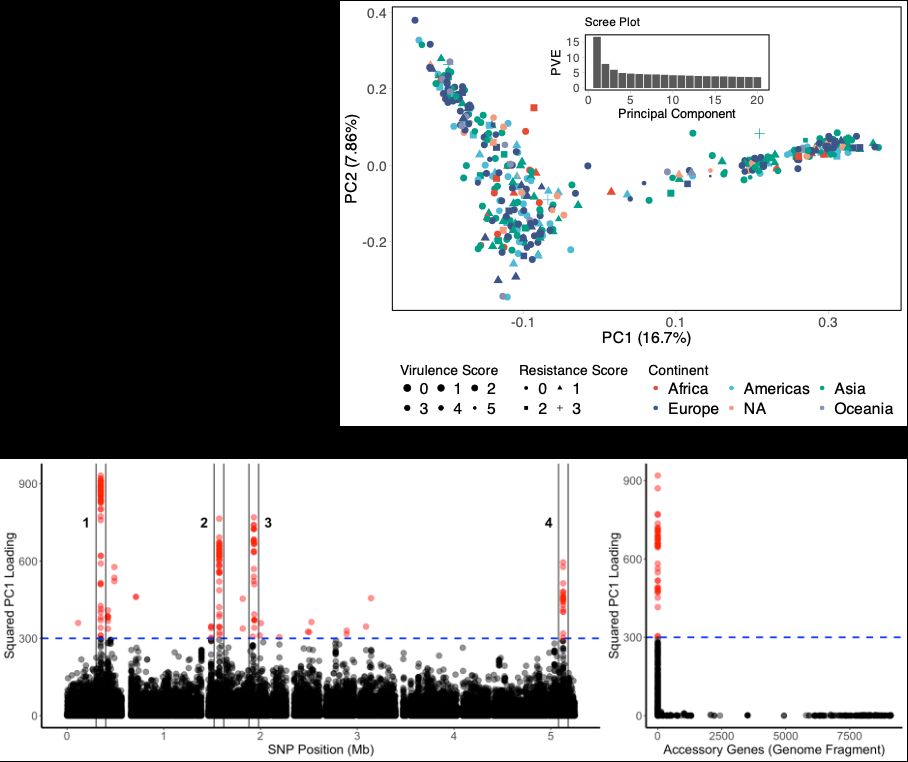
We think that this structure is due to a genome-wide component of variation, which can be captured for example using Principal Components Analysis (PCA). A handful of loci in the genome contribute disproportionately to PC1 values, these loci are the most different between different ends of the tree.
27.01.2025 08:34 — 👍 0 🔁 0 💬 1 📌 0
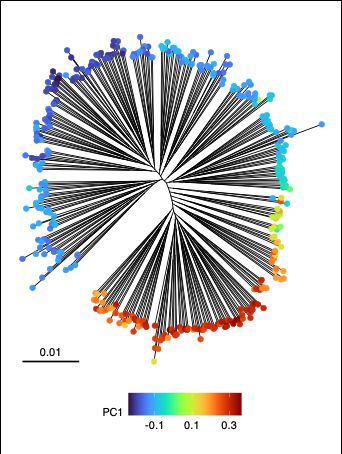
Have you never noticed the "backbone" in a tree of Klebsiella pneumoniae genomes? Did you ever wonder what might be causing it? If so, very interested to hear what you thought.
27.01.2025 08:34 — 👍 0 🔁 0 💬 1 📌 0
Professor of Life Sciences & Director Global Health Institute @EPFL_en. Passionate about science 🤩 Views are my own.
Associate Professor, MIT
Still thinking about the 10^9 mutations generated in your microbiome today.
Website: http://lieberman.science
School of Microbiology and APC Microbiome Ireland, University College Cork 🇮🇪
Microbial Ecology | Microbial Evolution | Gut Microbiome
Researcher at Uni Cambridge working on bacterial evolution and AI
Ecology and evolution in microbes. Assistant Prof. @ LIU Post, native NYer, foodie.
Assistant prof @ UofT, associated scientist @ Helmholtz Institute for RNA-based Infection Research. Pathogen systems biology / informatics / functional genomics. Coastal New Hampshirite. Drink Moxie.
Scientific coordinator & infection biologist @Sharmalab.bsky.social | RNA and infection | Deep-sequencing & microfluidics approaches to bacterial pathogenesis | Campylobacter & Helicobacter
Postdoctoral researcher - UMR8261 Expression Génétique Microbienne in Condon's lab.
Interested in post-transcriptionnal regulations, -omics, regulatory RNA, RNases, and sporulating bacteria 🦠
Alumni from Tine Arnvig's lab and David Hot's lab.
Postdoctoral fellow at NIH studying cell biology of coccoid bacteria.
Views my own.
He/him/Él 🏳️🌈
Molecular Principles of RNA Phages | Junior Professor @uni-wuerzburg.de | Group Leader @helmholtz-hiri.bsky.social
Interested in microbial ecology & evolution. Views are only my own. (he/him)
🎓: https://scholar.google.com/citations?hl=en&user=OBPpZq4AAAAJ&view_op=list_works&sortby=pubdate
👨💻: https://github.com/raufs
Macrobe qui aime les microbes
http://www.shapirolab.ca/
Microbiologist, NERC Independent Research Fellow, STEM ambassador
Vibrio | dormant bacteria | seafood | marine environments | Burkholderia
Postdoc at GenPhySE (INRAE Toulouse) on coupling quantitative genetics and population genetics methods for GWAS and genomic prediction
🌱 PhD student at Weigelworld @plantevolution.bksy.social | Graph pangenome | Population genetics
Bacterial RNA Biologist, Group Leader and Professor at Chinese Academy of Sciences, Shanghai Institute of Materia Medica (Formerly Institut Pasteur of Shanghai). Homepage: https://yanjiechao.wordpress.com
🇵🇹 Staff Scientist @probstlab.bsky.social
(@unidue.bsky.social - 🇩🇪). Microbial genomics in the One Health context, biogeochemistry of cave microbiomes, alga-microbe symbioses.
Professor of Genetics at U of Leicester. Genome structural variation.
"Anyone with gumption and a sharp mind will take the measure of two things: what's said and what's done." Views my own.
Professor of Algorithmic and Microbial Genomics at the University of Bath (UK). Pangenomes, drug resistance (esp TB), data structures for DNA search, plasmid evolution, global microbial surveillance. Open Data, reproducibility




















