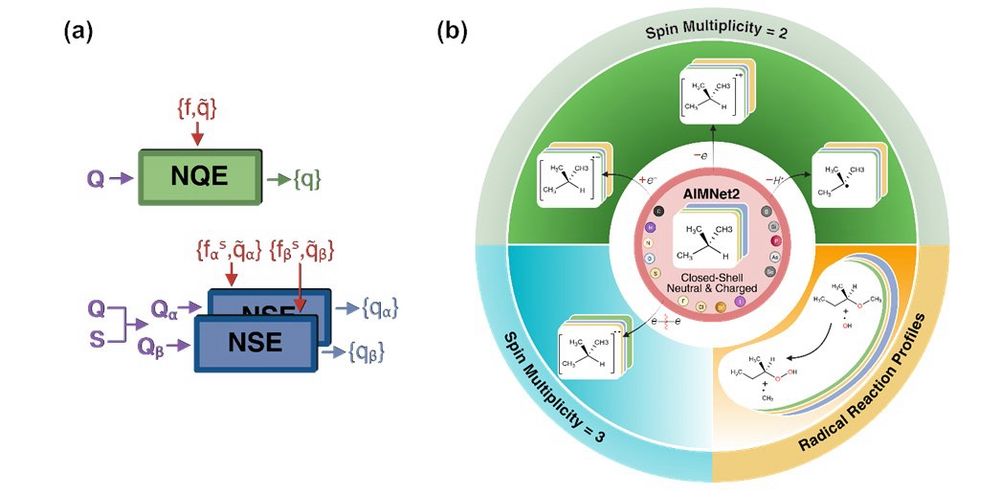
Most of MLIPs dont distinguish between spin states, making them unsuitable for open-shell chemistry. We present AIMNet2-NSE (Neural Spin-charge Equilibration), MLIP that incorporates spin-charge equilibration for systems with arbitrary charge and spin. #compchem #skychem
chemrxiv.org/engage/chemr...
04.08.2025 16:47 — 👍 23 🔁 3 💬 5 📌 0
Woohoo!
28.07.2025 16:01 — 👍 3 🔁 0 💬 0 📌 0
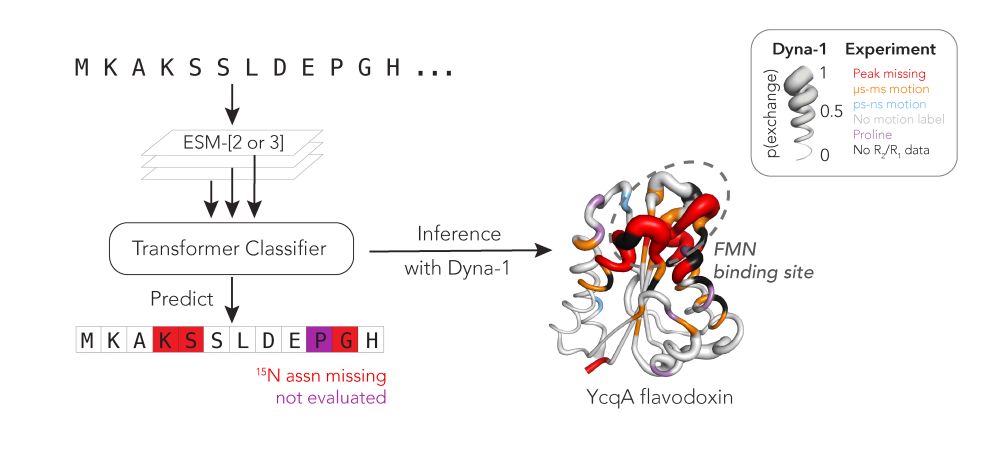
Protein function often depends on protein dynamics. To design proteins that function like natural ones, how do we predict their dynamics?
@hkws.bsky.social and I are thrilled to share the first big, experimental datasets on protein dynamics and our new model: Dyna-1!
🧵
20.03.2025 15:02 — 👍 102 🔁 38 💬 5 📌 5
Nooooooooo.... 😲
08.07.2025 22:04 — 👍 4 🔁 0 💬 0 📌 0
NIH funding supporting the HMMER and Infernal software projects has been terminated. NIH states that our work, as well as all other federally funded research at Harvard, is of no benefit to the US.
22.05.2025 12:42 — 👍 287 🔁 232 💬 37 📌 46
Our new preprint PharmacoForge: Pharmacophore Generation with Diffusion Models is out now! PharmacoForge quickly generates pharmacophores for a given protein pocket that identify key binding features and find useful compounds in a pharmacophore search. Check it out! 🧪 doi.org/10.26434/che...
27.05.2025 19:11 — 👍 21 🔁 9 💬 1 📌 0


Was proud and honored to hood Dr. Drew McNutt at the Pitt School of Medicine Diploma Ceremony. Here we are rocking both the blue and gold and tartan colors representing our joint Pitt-CMU CompBio PhD program. Congratulations to Drew and the other @cmupittcompbio.bsky.social graduates!
19.05.2025 13:55 — 👍 19 🔁 3 💬 0 📌 0
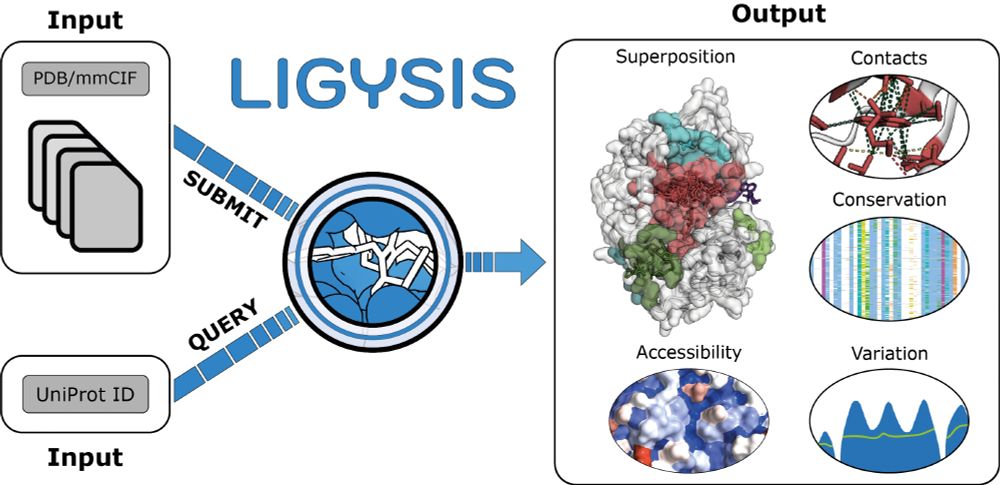
I am #exhilarated to share that our new paper "LIGYSIS-web: a resource for the analysis of protein-ligand binding sites" is now published in @narjournal.bsky.social! After almost two years of development and more than 600 commits, LIGYSIS-web is out!
📜 : tinyurl.com/utges-LIGYSI...
19.05.2025 10:37 — 👍 27 🔁 11 💬 2 📌 0
Job Details
We're recruiting a 3-year postdoc for the Novo Nordisk - Oxford Fellowship programme!
Develop machine learning approaches for fragment library design and experimental optimisation
With @fergusimrie.bsky.social and Charlotte Deane
Job advert: shorturl.at/3l47e
Further details: shorturl.at/u4UkK
08.04.2025 15:44 — 👍 4 🔁 5 💬 0 📌 0
Everything is easy in 2D.. - @franknoe.bsky.social
31.03.2025 17:03 — 👍 3 🔁 0 💬 0 📌 0

Interested in generative modeling and pharmacophores search for SBDD? Check out our talks at #ACSSpring2025
25.03.2025 15:15 — 👍 4 🔁 1 💬 0 📌 0
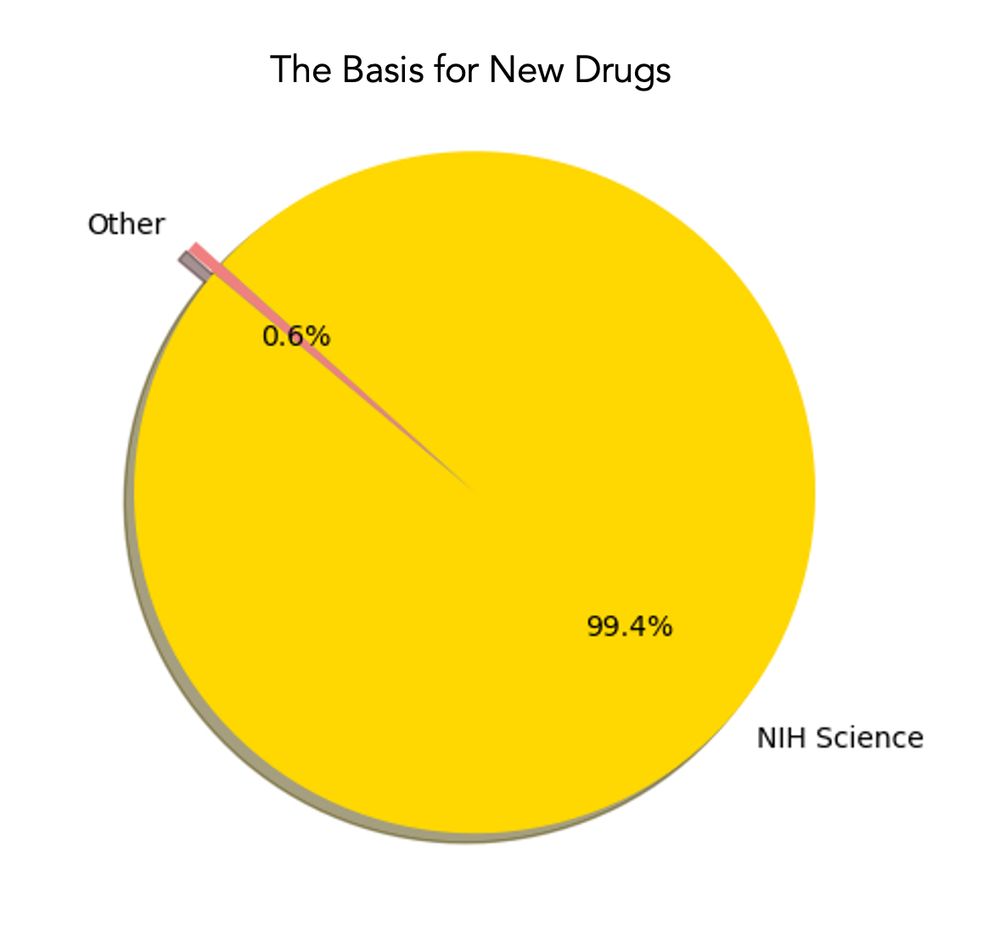
graph of NIH basisfor new drugs
A pie graph worth keeping in mind as the NIH budget plummets jamanetwork.com/journals/jam... for 356 new FDA drugs approved
23.03.2025 16:17 — 👍 4062 🔁 1668 💬 63 📌 86
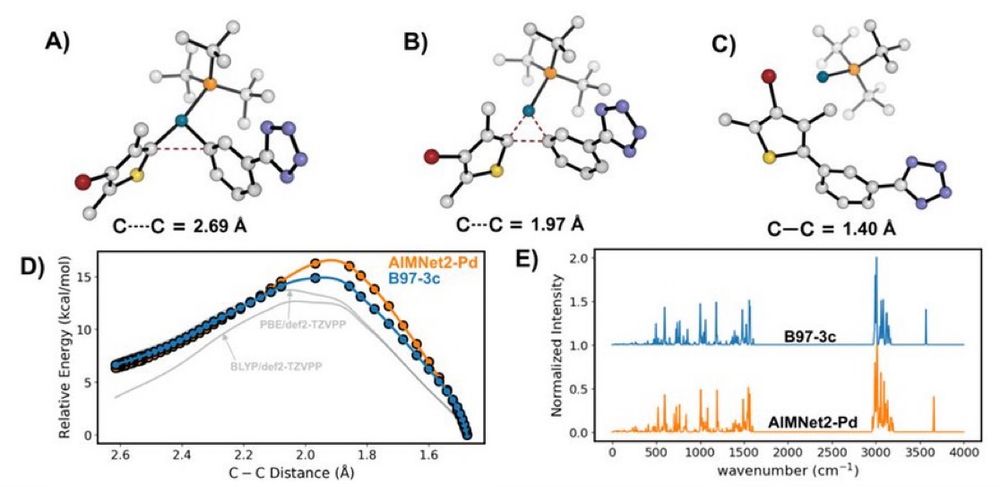
Transferable MachineLearning Interatomic Potential for Pd-Catalyzed Cross-Coupling Reactions
Our latest ChemRxiv preprint, "Transferable #MachineLearning Interatomic Potential for Pd-Catalyzed Cross-Coupling Reactions" Collaboration with @nsf-ccas.bsky.social @gabegomes.bsky.social @bobbypaton.bsky.social chemrxiv.org/engage/chemr... #compchem #chemsky
18.03.2025 14:12 — 👍 57 🔁 9 💬 4 📌 3
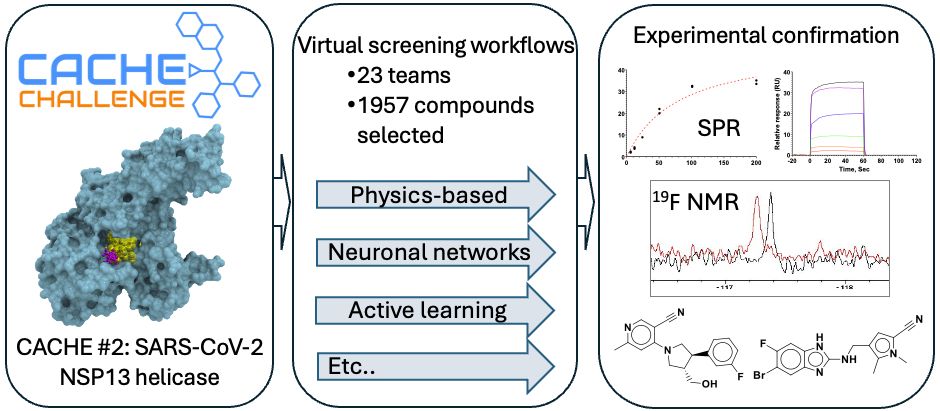
The CACHE #2 preprint is now online: bit.ly/3DyCmHN Active learning and fragment growing delivered confirmed hits. A citizen scientist using the Fold-it gaming interface designed the top compound! Kudos to Sasha and Madhushika @thesgc.bsky.social at the bench. @conscience-network.bsky.social
10.03.2025 21:40 — 👍 7 🔁 7 💬 0 📌 2


@standupforscience.bsky.social
07.03.2025 16:56 — 👍 7 🔁 1 💬 0 📌 0

Andrew McNutt on a sola climbing trip in New York

Andrew McNutt poses for a photo with other students in the CPCB program
Andrew McNutt, '24 CPCB graduate, is ready for life's next adventure. Before he begins his career in drug discovery, he's taking a well-deserved break to hike the Appalachian Trail from Georgie to Maine.
Read more: tinyurl.com/AndrewMcNutt
17.02.2025 16:11 — 👍 4 🔁 1 💬 0 📌 0

Welcome to the Bluesky account for Stand Up for Science 2025!
Keep an eye on this space for updates, event information, and ways to get involved. We can't wait to see everyone #standupforscience2025 on March 7th, both in DC and locations nationwide!
#scienceforall #sciencenotsilence
12.02.2025 17:04 — 👍 11587 🔁 5520 💬 292 📌 679
I’ll have to read the paper, but that’s a bizarre take given the graph. The issue isn’t the amount of data but the ability to generalize from it - which absolutely requires more innovative algorithms.
08.02.2025 15:06 — 👍 2 🔁 0 💬 1 📌 0

AlphaFold can predict protein, DNA, or RNA complexes.. BUT is it easy to determine the reliable interactions?🤔
➡️ AlphaBridge will identify the most confident interactions, making your analysis easier!
🌐Try it here: alpha-bridge.eu
👀Read more: www.biorxiv.org/content/10.1...
05.11.2024 00:18 — 👍 119 🔁 28 💬 6 📌 1
NIH council meeting (NIDCR)scheduled for today, where our R01 grant (3rd percentile) was supposed to be discussed, was postponed indefinitely as a consequence of Trump's executive order which ordered pausing of all communications from all federal agencies. Not good.
22.01.2025 17:18 — 👍 163 🔁 82 💬 18 📌 16
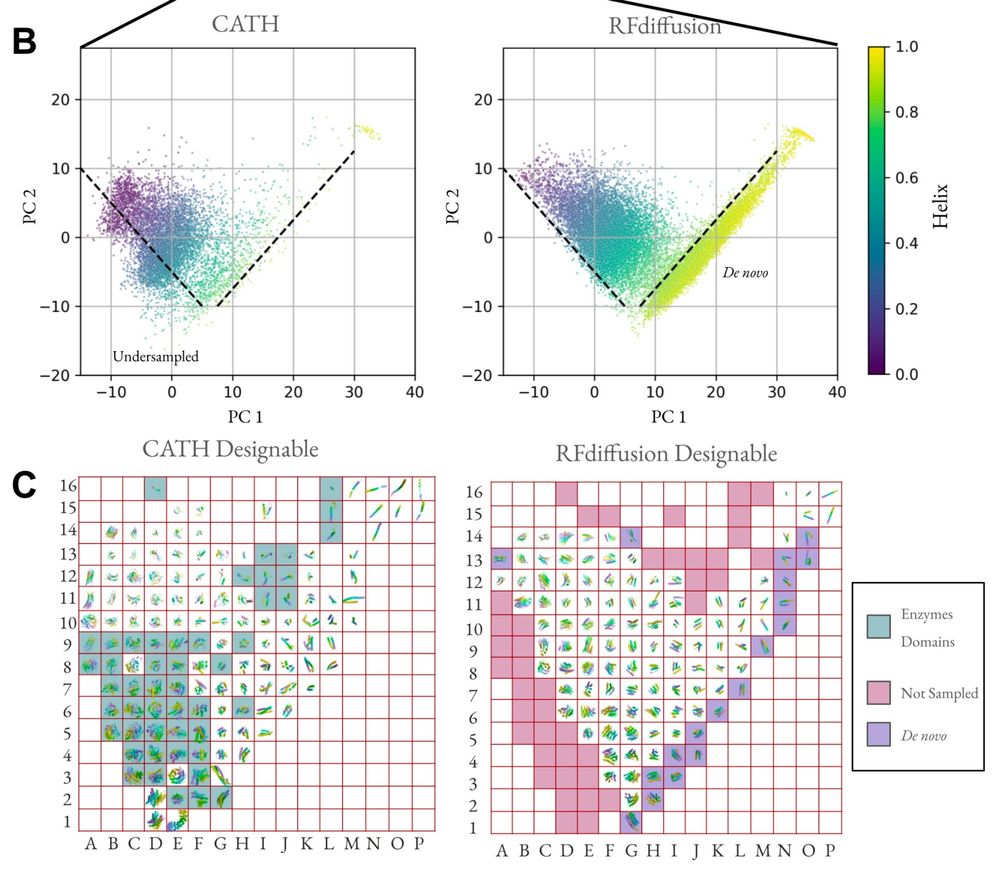
Protein structure embeddings reveal undersampled and de novo structure space. (B) First two principal components of mean-pooled ESM3 embeddings colored by helix content determined by DSSP. The indicated dashed guide lines denote visual boundaries of native structure space not sampled (Undersampled) and novel regions of protein structure for this space only observed in samples but not in native structures (De novo). (C) Rasterized visualization of panel B with 16 equally spaced grid squares in each principal component axis. A representative structure from each grid was chosen at random. Empty grid squares indicate the absence of any structure in the enclosed region. De novo alpha helices are shaded along the lower-right diagonal and the structures from CATH which do not have corresponding structures in the samples are shaded along the left and top rims. The structures are displayed in CATH raster plot are given in the Supplementary Information.
Protein backbone diffusion models consistently undersample catalytically important motifs, and oversample idealized helices, raising questions about the appropriateness of these methods in designing starting points for enzyme design & evolution. From www.biorxiv.org/content/10.1...
19.01.2025 16:40 — 👍 86 🔁 16 💬 2 📌 0
OPIG
Oxford Protein Informatics Group
OPIG is now on Bluesky!
Follow us for updates about the group's latest work, web app updates, and more.
opig.stats.ox.ac.uk
15.01.2025 13:53 — 👍 11 🔁 6 💬 0 📌 0
If you want to learn more about BioEmu, join Sarah Lewis and Michael Gastegger in @hannes-stark.bsky.social ‘s online seminar today.
bsky.app/profile/hann...
13.01.2025 08:10 — 👍 19 🔁 5 💬 1 📌 1
Rishal’s colab notebook will suggest pharmacophores given a receptor structure that can be directly imported into Pharmit for screening. Check it out!
03.01.2025 20:18 — 👍 7 🔁 2 💬 0 📌 0

2024 Year-In-Review
While it’s not strictly the end of 2024, I wanted to draft this early. Obviously a highlight is the release of 1.99 in time for Ubuntu LTS (24.04) … a summer of great coding / rendering, leading to t...
I wrote up a 2024 Year in Review for @avogadro.cc 🧪⚗️ #opensource #chemsky
In short, the community is amazing. Significant translations into 17 languages including 🇭🇷🇫🇷🇭🇺🇰🇷🇷🇴🇪🇸Tamil, 🇺🇦🇨🇳🇬🇪🇩🇪🇯🇵🇵🇹🇧🇷🇷🇸🇹🇷
.. and of course dozens of features and bug fixes discuss.avogadro.cc/t/2024-year-...
30.12.2024 15:58 — 👍 15 🔁 5 💬 1 📌 0
I get it, and I can't adequately express how grateful I am for the PLINDER resource, but if you label something the test set, then people will `splits=[ "test"]` and use it uncritically. If the MLSB test set is the right one, why not label it "test" and have a larger "test-with-duplicates"?
24.12.2024 17:21 — 👍 3 🔁 0 💬 1 📌 0
These ligands make up 33% of the v2 plinder test set (shown with counts):
MG 94
NAG 72
NAG-NAG 64
ZN 44
MN 32
BMP 30
HEM 24
EDO 18
SO4 17
PO4 16
ADP 15
GOL 13
ANP 13
CA 13
FMN 11
24.12.2024 17:06 — 👍 6 🔁 0 💬 2 📌 0
Computational biologist surviving the chaos of grad school and parenting—fueled by coffee.
Virologist & Professor, Pitt Dept. of Microbiology & Molecular Genetics and former Director of @Pitt-PMI.bsky.social
Seattle native living in Pittsburgh.
Website: http://ambrose-lab.com
#MolecularSimulations | Asst. Prof. Tel Aviv University | PostDoc: Rothschild Fellow, Parrinello group, ETH Zurich | PhD: Adams Fellow, w/Benny Gerber, Hebrew University | #newPI
Computational Biologist @ CMU
NHGRI K99 Awardee, Postdoc in the Noble Lab, UW. Ph.D. in CS and Comp bio, Biociphers Lab, Penn. Pronouns: she/her
Comp bio lab @ Pitt. Makes & applies tools for pharm & basic-science research. Cheminformatics, MD simulations, visualization, computer docking.
I'm just trying to get through life alive
Because Science is for everyone! 🧪🌎
Learn more ⬇️
http://linktr.ee/standupforscience
👯sister org: @sciforgood.bsky.social
Professor, Carnegie Mellon University; computational biophysicist and theoretical chemist
SNSF Ambizione Project Leader
Looking at tiny frozen mitochondria with big microscopes 🔬 cryoEM/cryoET
Green organisms 🌿 & evolution 🦠
Biozentrum, hosted in the Engel Lab, CH
🇫🇷 -> 🇩🇪 -> 🇨🇭
Website: https://www.waltzresearch.com/
I live in Switzerland now. @cellarchlab.com at the @biozentrum.bsky.social
Running a Structural Biology and Protein Production group at a Boston biotech. Previously worked in rare disease, but focusing on drug discovery in precision oncology the last few years. Crystallography with a splash of CryoEM and NMR. Opinions are my own.
We use structure-guided engineering to design biologics & immunotherapies.
Associate Professor of Immunology,
Director - Center for Antibody & Protein Engineering (CAPE) @ Moffitt Cancer Center
www.thelucalab.org
Photosynthesis and cryoEM at Biozentrum, Basel.
Cellular Structural Biology Wellcome DPhil Student at The University of Oxford.
Currently working in Peijun Zhang’s lab at The Oxford Division of Structural Biology.
Passionate about biomedical animation and science communication: Batisio.co.uk
Science, NMR Spectroscopy, proteins/lipids, dynamics in Gif-sur-Yvette, but also music, guitar, ha brezhoneg...
Postdoc in the Banfield lab @ John Innes Centre | PhD in the Williams lab @ ANU | Plant immune receptor engineering 🌾 | Fungal effectors 🍄 | Structural biology 💎 | 🇦🇺
Cryo-EM @Prof Daniel Wilson Lab, Uni of Hamburg, Germany
Single particle cryo-EM, cryo-electron tomography
BS RIT | PhD UMich - Janet Smith Lab | Postdoc MIT - Imperiali & Kiessling Labs |
Ruth L. Kirschstein Fellow | Senior Research Fellow @ Biogen
Membrane proteins, glycobiology, structural biology, bikes, dogs, bills mafia



















