This is a great read - it nails a whole bunch of ways in which LLMs simply don’t ‘think’, let alone at a ‘PhD level’.
09.08.2025 21:06 — 👍 13 🔁 6 💬 1 📌 1
But perhaps most critically, it’s (imo) irresponsible to attribute this to *science* as a whole; suggesting it’s a monolith when things like the image duplication retractions are pretty limited to a unique corner of science under sleuth spotlights.
06.08.2025 12:38 — 👍 70 🔁 4 💬 2 📌 0

Logo for the Genomic History Inference Strategies Tournament.
If you're new to demographic history inference from population genomics, try this webapp I created to illustrate how dadi fits bottleneck models to site frequency spectra: ryangutenkunst-dadi-two-epoch.hf.space . It even outputs files for submitting to the GHIST competition! ghi.st
05.08.2025 18:48 — 👍 37 🔁 21 💬 1 📌 2
🧬🏺Calling everyone interested in the intersection of archaeological theory and ancient DNA!
Thrilled to be co-organizing a session for #TRAC2025 with @ezgimou.bsky.social
The virtual conference will be held October 22 - 24 and the call for abstracts is now open. Come join us!
05.08.2025 18:43 — 👍 11 🔁 3 💬 1 📌 0

🧪 🏺
Excited to co-organize a session with @mootspoints.bsky.social at #TRAC2025!
🧬 Integrating Ancient DNA with Archaeological Theory and Practice
We welcome papers on mobility, migration, & identity in the Roman world and beyond.
📢 Call for abstracts now open! Please spread the word.
#aDNA #science
05.08.2025 13:49 — 👍 16 🔁 7 💬 1 📌 2
Check out our paper in Evolution Letters!
We used D. melanogaster pigmentation as a focal trait to explore parallelism in phenotypic and genomic responses to environmental change - read more at the link below 👇
31.07.2025 21:32 — 👍 52 🔁 22 💬 2 📌 1
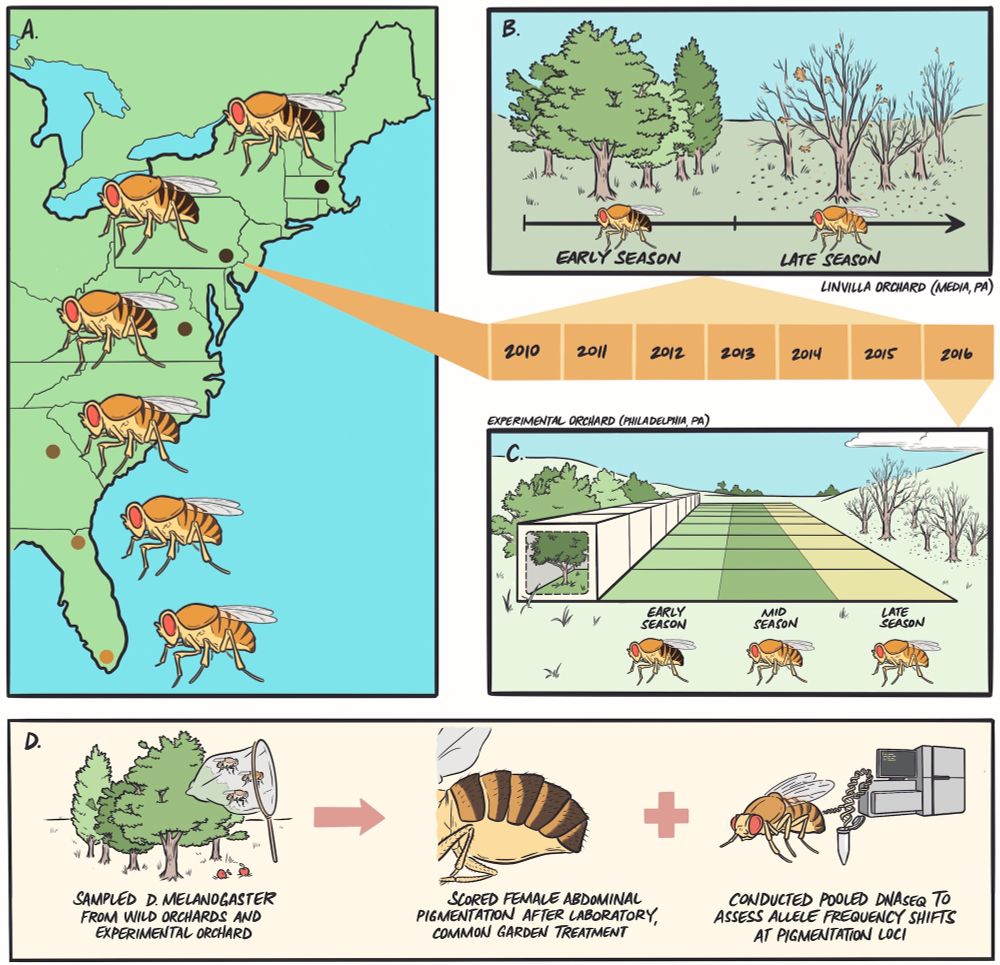
Figure showing the experimental overview as a graphic. Remaining alt text taken from the figure caption in the paper: (A) we sampled flies from six wild orchard populations ranging from Homestead, FL, to Lancaster, MA, and established isofemale lines in the laboratory. (B) We returned to a focal orchard in Media, PA, at early- and late-season timepoints and collected flies to capture evolutionary patterns following winter and summer conditions. (C) We then seeded outdoor mesocosms (N = 9) with an outbred population originating from early-season collections in Media, PA, and sampled flies at the end of summer (mid-season) and fall (late-season) to determine if seasonal patterns are recapitulated in experimental populations controlled for migration, drift, and cryptic population structure. (D) Across each wild or experimental context, we sampled flies, established lines in the lab, completed common garden treatment to remove environmental effects, and scored females for abdominal pigmentation. We also conducted pooled DNA sequencing on additional flies sampled from each population to map genomic patterns for candidate pigmentation SNPs.
How predictably does complex trait adaptation proceed over space and time in wild populations?
doi.org/10.1093/evle...
Now in @evolletters.bsky.social by @skylerberardi.bsky.social, @paulrschmidt.bsky.social et al.
📷: Dr. Rush Dhillon
31.07.2025 18:35 — 👍 24 🔁 11 💬 0 📌 1
Home - ProbGen 2026
Your Site Description
The 2026 Probabilistic Modeling in Genomics (ProbGen) meeting will be held at UC Berkeley, March 25-28, 2026. We have an amazing list of keynote speakers and session chairs:
probgen2026.github.io
Please help spread the news.
06.06.2025 17:52 — 👍 64 🔁 35 💬 2 📌 0
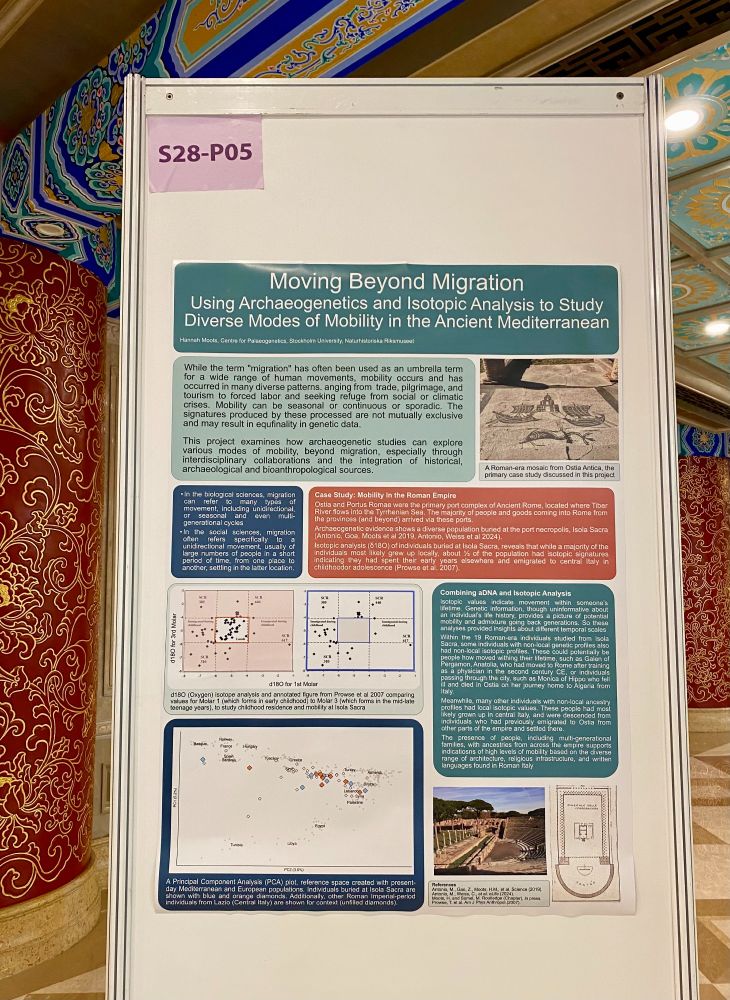
Come check out my poster this afternoon! On the language we use to talk mobility in ancient DNA studies #SMBE2025
22.07.2025 04:58 — 👍 46 🔁 7 💬 5 📌 0
Hence, this highlights the fundamental difference btw genomic and family h2. The former is considering millions of years of mutation while the latter is looking at everything but mutations over a very short period of time.
20.07.2025 07:26 — 👍 5 🔁 2 💬 1 📌 0
I can now finally speak out loud my honest thoughts on missing heritability or the genomics/family gap in h2 estimates.
Based on new paper the *issue* is a feature and not a bug.
TLDR: SNP h2 only captures mutational variance and family h2 captures everything but mutation.
1/n
20.07.2025 07:04 — 👍 16 🔁 3 💬 1 📌 1
New preprint comparing aDNA preservation and recovery rates during extraction from all three ossicles and the petrous. Congrats @compevohumang.bsky.social!
20.07.2025 09:40 — 👍 10 🔁 5 💬 0 📌 0
Join us at the Royal Society in London 14 November for a day on Ancient genomes, human biology and medicine!
Many slots are reserved for talks from submitted abstracts related to the topic, alongside speakers Svante Pääbo, Priya Moorjani, @mathiesoniain.bsky.social and Lluis Quintana-Murci.
14.04.2025 12:23 — 👍 9 🔁 6 💬 0 📌 1
Nifty preprint led by @mathiesoniain.bsky.social comparing femur length as a proxy of stature to genetic variants linked to height today. Many key insights, but main one for me is that variants responsible for lactase persistence are strongly associated with height in ancient but not modern people.
18.07.2025 07:28 — 👍 30 🔁 7 💬 2 📌 1
My talk from this year's probgen is finally out.
Where do linear mixed models and random effects come from?
They emerge from *mutations* on Ancestral Recombination Graphs.
19.07.2025 01:04 — 👍 48 🔁 22 💬 2 📌 2
A difficulty in scrutinising ancient DNA from the outside is that the original methods are discussed in arcane terms in separate papers cast across the innumerable winds of scientific publishing. Here are 2 papers which attempt to discuss collections of common methods with plainer language.
18.07.2025 10:46 — 👍 24 🔁 7 💬 0 📌 0
It started with a meeting between @mp-williams.bsky.social) and Robbi Davidson during the last @isbarchaeology.bsky.social conference (Tartu, 2023). I remember them sitting at the bar after lunch, engrossed in their discussion—that moment was even captured by the event photographer 🧵1/n
03.07.2025 23:52 — 👍 6 🔁 1 💬 1 📌 0

Get excited for ISBA11! The conference program is now online!
www.isba11.com/programme
#ISBA11 #biomoleculararchaeology #Archaeology #Archaeology #archaeologist
@isbarchaeology.bsky.social @archaeobiomics.bsky.social
01.07.2025 11:41 — 👍 16 🔁 9 💬 0 📌 1
👇 Make sure to register for our webinar happening next week!
11.06.2025 10:33 — 👍 2 🔁 1 💬 0 📌 0

Professur für Archäologie des antiken Mittelmeerraumes und seiner Umwelt (W2)
Open position at @unileipzig.bsky.social for professor of archaeology of the antique Mediterranean! www.uni-leipzig.de/stellenaussc...
03.06.2025 10:48 — 👍 3 🔁 3 💬 0 📌 0
I have an opportunity to hire a staff scientist for my lab. Looking for someone with outstanding skillset in ML/statistics, genomics applications; interest in mentoring, strong publication record, PD experience required.
Email CV to me+cc my assistant (see 'contact' on my website). Ad to follow.
01.06.2025 15:33 — 👍 82 🔁 108 💬 3 📌 3
🥳 We're back with a new edition of our HAAM-radio webinar series! Come and be part of the development from a reporting scheme for aDNA samples & seqdata!
#aDNA #workshop #humanDNA #sequencing
13.05.2025 13:28 — 👍 6 🔁 5 💬 0 📌 0
PhD candidate | JGU Mainz | Palaeogenetics Group | Palaeo-epigenomics | aDNA | 🇪🇬
PhD Student at UMich Statistics.
The account mostly trashes about urban planning and infrastructure.
Probability, Statistics, and Evolutionary Biology.
https://hanbin973.github.io
Postdoctoral Researcher at the University of Oxford, and Ludwig Maximilian University of Munich • Ancient DNA, Domestication, Dogs • 🇳🇿
Writer, journalist. Science, health. Pandemics, animals. Birder, photographer. Many words, some awards. AN IMMENSE WORLD, I CONTAIN MULTITUDES. Married to Liz Neeley, parent to Typo. he/him
📷 Canon R6mkii + RF 800mm
Edyong.me
AI and cognitive science, Founder and CEO (Geometric Intelligence, acquired by Uber). 8 books including Guitar Zero, Rebooting AI and Taming Silicon Valley.
Newsletter (50k subscribers): garymarcus.substack.com
The first edition of the International Conference on Palaeogenomics (ICP2026) will be in Stockholm, 23–26 June 2026.
More details soon!
https://icp2026.palaeogenomics.org/
Associate Professor
DFCI & HMS
Postdoctoral researcher at the University of Wisconsin studying South American genetic history 🇨🇴🇪🇸
Into: 🐱🤸🏾♀️🧘🏾🥑🍺🎪
Member of the SMBE IDEA Taskforce & HAAM community 🌿
https://t.co/S7RLrxWE20
PhD student in the Durbin group; Uni of Cambridge. Currently, visiting researcher in the Reich Lab, Harvard University.
Human evolutionary genetics, University of Cambridge; Darwin College. 🇮🇪
Ancient DNA Lab at the Center for New Technologies, University of Warsaw. Working on 🦇🐿️🐀🦌🐻🫎🐐🐈⬛🧔♂️🦇🐎 and more!
http://adna.cent.uw.edu.pl/
Archaeogeneticist - Postdoctoral Research Associate at Brown University
Population genetics | ancient DNA | human health
Junior Research Fellow at Magdalene College, Cambridge
Australian Centre for Ancient DNA
Ad Astra-Assist. Prof at UCD. Young Academy Ireland. https://t.co/ZQ4XYYxM7H
Ancient DNA, goats, sheep (begrudgingly), pathogens, climate, MTG, The Traitors.
I watch too many films: http://letterboxd.com/GingerHowley
He is openly gay (citation needed)
Banting & Killam Postdoc @ ADaPT lab, UBC
archaeogenomics, fisheries, marine historical ecology | herring 🐟
National Geographic Explorer
views my own
Genetics researcher @ ANU
Postdoc in evolutionary genomics of polyploids at RBG Kew | science & society | opinions mine
Mizzou, Fulbright Belgium, and Michigan State alum. Here because of medicaid, public universities, and pell grants
https://kevinabird.github.io/