Overjoyed to finally have our isopeptide bond paper out! If you're interested in the types of proteins and organisms that use a cool intramolecular covalent bond, check it out:
bit.ly/4od3ux8
Plenty of future SynBio/EngBio applications planned! Thanks to all involved (character limits suck).
22.11.2025 05:33 — 👍 4 🔁 2 💬 1 📌 0
Phil Hinchliffe and Steve Burston, Delhi Kalwan, Jennifer de Jong, Fabio Parmeggiani and Paul Race
13.05.2025 16:51 — 👍 0 🔁 0 💬 0 📌 0
This project was made possible thanks to a collaboration between @bristoluni.bsky.social and @ebi.embl.org. A great thanks goes to @robbarringer.bsky.social, Ioannis Riziotis (Crick), Antonina Andreeva,
@alexbateman1.bsky.social and ...
13.05.2025 16:51 — 👍 0 🔁 0 💬 1 📌 0
⚠️⚠️⚠️Preprint alert⚠️⚠️⚠️
We mapped intramolecular isopeptide bonds across the AlphaFold database and found that they are widely distributed in microbial surface proteins, such as fibrillar adhesins or pilins, suggesting new targets for broad-spectrum antimicrobial strategies.
13.05.2025 16:43 — 👍 4 🔁 1 💬 1 📌 0
🙏 Special thanks to my amazing collaborators: Ioannis Riziotis, @robbarringer.bsky.social, Antonina Andreeva and to my supervisor @alexbateman1.bsky.social for their invaluable contributions to this work!
20.03.2025 10:54 — 👍 0 🔁 0 💬 0 📌 0

Google Colab
💻 Available as a Python package for easy integration into bioinformatics workflows, and accessible via Google Colab for everyone: colab.research.google.com/github/Franc...
20.03.2025 10:54 — 👍 1 🔁 0 💬 1 📌 0

🧬 Isopeptor enables reliable detection and geometry evaluation of these covalent links using a template-based strategy powered by pyJess.
🔍 The tool demonstrates a precision of 1.0 and recall of 0.947 in identifying incorrectly modelled isopeptide bonds in PDB structures.
20.03.2025 10:54 — 👍 0 🔁 0 💬 1 📌 0
Check out this article about my research at @embl.org where I focus on protein modelling and on the study of fibrillar adhesins. Recently, we developed a method to detect isopeptide bonds, a key stabilizing feature in bacterial proteins. Thanks @oanastroe.bsky.social for putting this together!👇 👇 👇
18.02.2025 18:43 — 👍 12 🔁 2 💬 0 📌 0
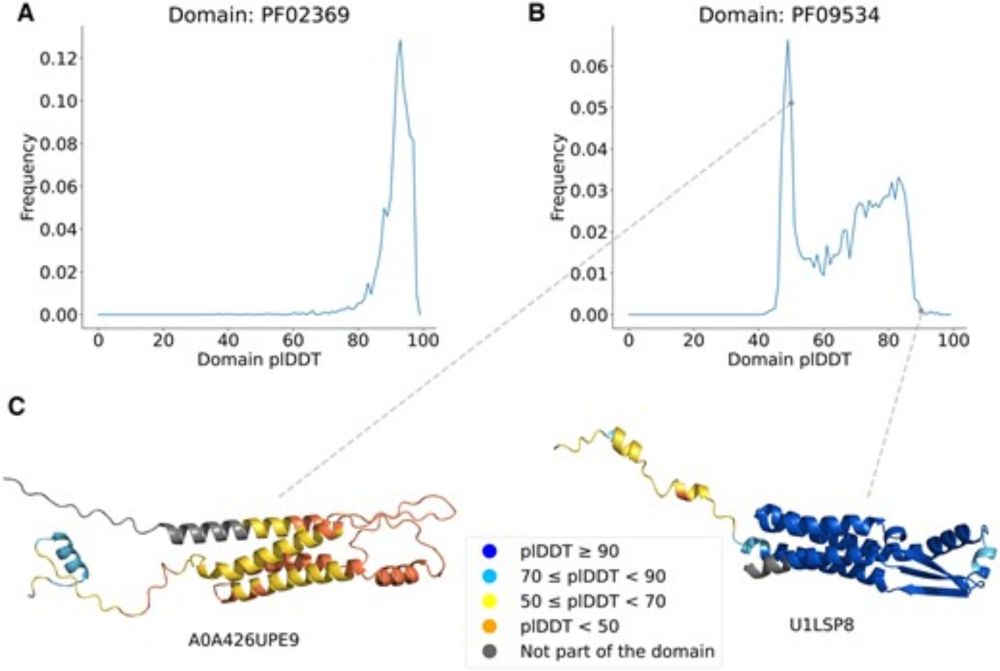
Protein structure prediction from the AlphaFold Database
Prediction confidence scores help #AlphaFold users gauge the reliability of protein structure predictions. 🖥️🧬
But things get more challenging for protein families.
A new method helps improve low confidence predictions within protein families.
academic.oup.com/bioinformati...
16.12.2024 09:28 — 👍 17 🔁 7 💬 0 📌 0
Thanks Gonzalo!
13.12.2024 11:44 — 👍 1 🔁 0 💬 0 📌 0
A special thanks goes to my supervisor @alexbateman1.bsky.social
and to @matthiasblum.bsky.social
for their support and guidance. 7/7
09.12.2024 13:57 — 👍 2 🔁 0 💬 0 📌 0
Our findings have important implications for improving structure predictions, especially for proteins from organisms with limited representation in sequence databases or for rapidly evolving taxa. 6/7
09.12.2024 13:57 — 👍 0 🔁 0 💬 1 📌 0

We show that using high plDDT models as templates can increase the speed of AlphaFold2 as implemented in ColabFold, potentially reducing computational costs and carbon footprint. 5/7
09.12.2024 13:57 — 👍 1 🔁 0 💬 1 📌 0

We introduced a novel "Best Pick" strategy that combines predictions made with and without multiple sequence alignment (MSA) information, selecting the model with the highest average plDDT. 4/7
09.12.2024 13:57 — 👍 0 🔁 0 💬 1 📌 0

This observation led us to explore whether low-confidence predictions could be improved using high-confidence templates from the same protein family. About one-third of low-confidence structures can be "rescued" to reasonable confidence levels using this method. 3/7
09.12.2024 13:57 — 👍 0 🔁 0 💬 1 📌 0

By observing the plDDT distribution within protein domain families, we noticed a certain degree of heterogeneity in the confidence of AlphaFold2 predictions. 2/7
09.12.2024 13:57 — 👍 1 🔁 0 💬 1 📌 0
Co-Creating Ireland's Public Involvement in Open Research Roadmap
ENGAGED is building a national roadmap to shape public involvement in open research in Ireland. We believe that research can and does play an important role in tackling societal challenges.
RNA Resources Project Leader at @ebi.embl.org RNAcentral, Rfam
The Open Bioinformatics Foundation (OBF) is a non-profit volunteer-run organization that supports #OpenSource #bioinformatics. Member projects: #BioPerl, #BioJava, #BioRuby and #Biopython. We run an annual meeting, the Bioinformatics Open Source Conference
I like to jump, jump, jump.
Yo mama so old, she invented wheels.
You know Etsy? Yeah, I've a store there: https://thebadappleid.etsy.com.
Associate Prof of BME at BU
Synthetic mechanobiology, single molecules, molecular structure and engineering, bio-organic chemistry, and general enthusiast of science, kindness, and understanding.
www.thengolab.com
Protein folding nerd, still a grad student (at my age? yes!!) UW IPD. She/any and AutAF
Ph.D. student at UT Southwestern Medical Center.
PhD student@MIT
https://sites.google.com/view/yehlincho/home
L’Institut du #Cancer AP-HP. Nord - Université Paris Cité acteur majeur de la cancérologie en Ile-de-France. 🔬🦠
https://institutducancer-hopitauxnord-u-paris.aphp.fr/
🧬 Structural Bioinformatics | 💊 AI/ML for Drug Discovery | Geometric DL
🔬 @iocbprague.bsky.social, prev. PhD @cusbg.bsky.social @mff.unikarlova.cuni.cz
Love epidermal stem cells and lipids. EMBL Predoc | Erasmus Master Mundus IMIM
Mathematician - Interested in ML, Geometry, Optimization, Time Series https://scholar.google.com/citations?user=vIJxhvcAAAAJ&hl=en
https://github.com/dnguyend/
🔬 Advancing RNA, DNA, Exosomes & Protein Research
🌎 Trusted in 120+ Countries Worldwide
👇🏽 Empowering Scientists with Innovative Solutions
🔗 linktr.ee/norgenblue
PhD candidate at EMBL-EBI and the University of Cambridge (Uhlmann Group). Turning bioimages into numbers and finding the stories they hide. Loves math, climbing, and anime
Assistant Professor @ucf.bsky.social
https://medvedev-lab.cs.ucf.edu/
Alumni @utswtxid.bsky.social
Computational Biology | Bionformatics | Cancer Biology
Group leader, Royal Society Industry Fellow,
proteins, RNA, biotechnology, structural biology, biophysics, protein engineering, scaffolds, biosensing, nanopores, etc
University of (Olde) York, UK 🇬🇧🇨🇦🇫🇷🇪🇺
LinkedIn: mjplevin
PhD from Open University School of Planetary Sciences researching Mars's climate, focusing on Lyot Crater, after my Physics BSc. Previously did 3 degrees in Roman archaeology, one in languages and a law degree. Multinational upbringing. 🇯🇪🇨🇦🇩🇪