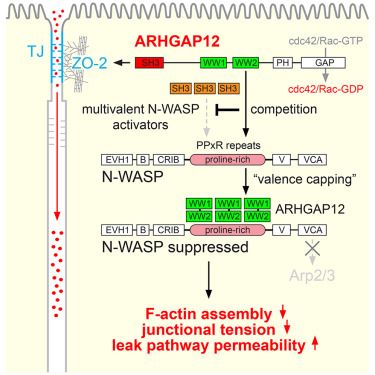
We have some news! We find that the post-synaptic Homer proteins coordinate YAP and Wnt signaling in epithelial cells, downstream of the Crumbs polarity complex. They do so by sequestering YAP and beta-catenin in biomolecular condensates, and by suppressing a FRYL/NDR complex. Check it out!
25.07.2025 13:23 — 👍 5 🔁 2 💬 0 📌 0

Intracellular mechanics and organelle mechanobiology
Mechanobiology is an interdisciplinary field that emerges at the cross-section of biology, physics and engineering. It aims to understand how living cells, tissues and animals sense and respond to me…
Announcing a new @embo.org Workshop on "Intracellular mechanics and organelle mechanobiology" that we are organizing with Michael Krieg and Verena Ruprecht at @icfo.eu (Barcelona) on 16-20 February, 2026. Please, repost and spread the word! 🙏 #EMBOmechanobio
meetings.embo.org/event/26-org...
04.07.2025 15:50 — 👍 56 🔁 36 💬 1 📌 1

EMBO Cell Polarity and Membrane Dynamics workshop is off to a good start - as always :) #EMBO
18.05.2025 14:58 — 👍 4 🔁 0 💬 0 📌 0
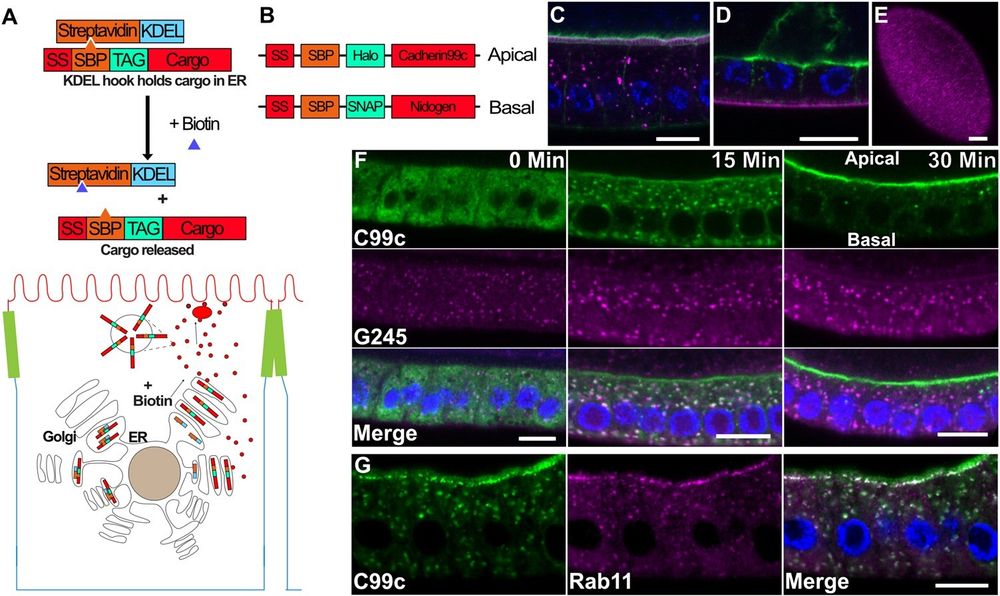
A) Diagram of the RUSH system. The Halo-tagged cargo is retained in the ER by the binding of the Streptavidin binding peptide (SBP) to Streptavidin (StrepA), which is fused to the ER retention signal, KDEL. The cargo is synchronously released upon the addition of biotin, which outcompetes SBP for binding to Streptavidin.B) Diagram of the tagged cargo constructs used in this study. C) Steady state expression of UAS-SBP-Halo-Cadherin99c (magenta) under the control of traffic jam-Gal4, showing its localization to the apical microvilli. Phalloidin staining of F-actin (green) labels the apical microvilli in the follicle cells and the oocyte. Scale bar 10 µm. D) Steady state expression of UAS-SBP-SNAP-Ndg (magenta) under the control of traffic jam-Gal4, showing its localization to the basement membrane. Actin is shown in green. Scale bar 10 µm. E) En face view of SBP-SNAP-Ndg in the basement membrane. Scale bar 10 µm. F) Time course of SBP-Halo-Cadherin99C trafficking in fixed samples. G) 25 min after release from the ER, Cad99c (green) localizes to subapical puncta that are labeled by Rab11 (magenta). Scale bar 10 µm. In all figures with a cross-section of the follicle cells, apical is toward the top of the image and basal toward the bottom.
How are specific cargos targeted to apical & basolateral domains within #EpithelialCells? This study uses a novel #vesicle tracking software "MSP-tracker" to show that the secretory pathway in #Drosophila follicle cells is unexpectedly spatially organized @plosbiology.org 🧪 plos.io/3EfJsBp
14.04.2025 11:58 — 👍 34 🔁 14 💬 0 📌 1
Entzugserscheinungsangst :)
17.04.2025 14:02 — 👍 2 🔁 1 💬 0 📌 0
Why am I not surprised…
17.03.2025 12:50 — 👍 2 🔁 0 💬 0 📌 0
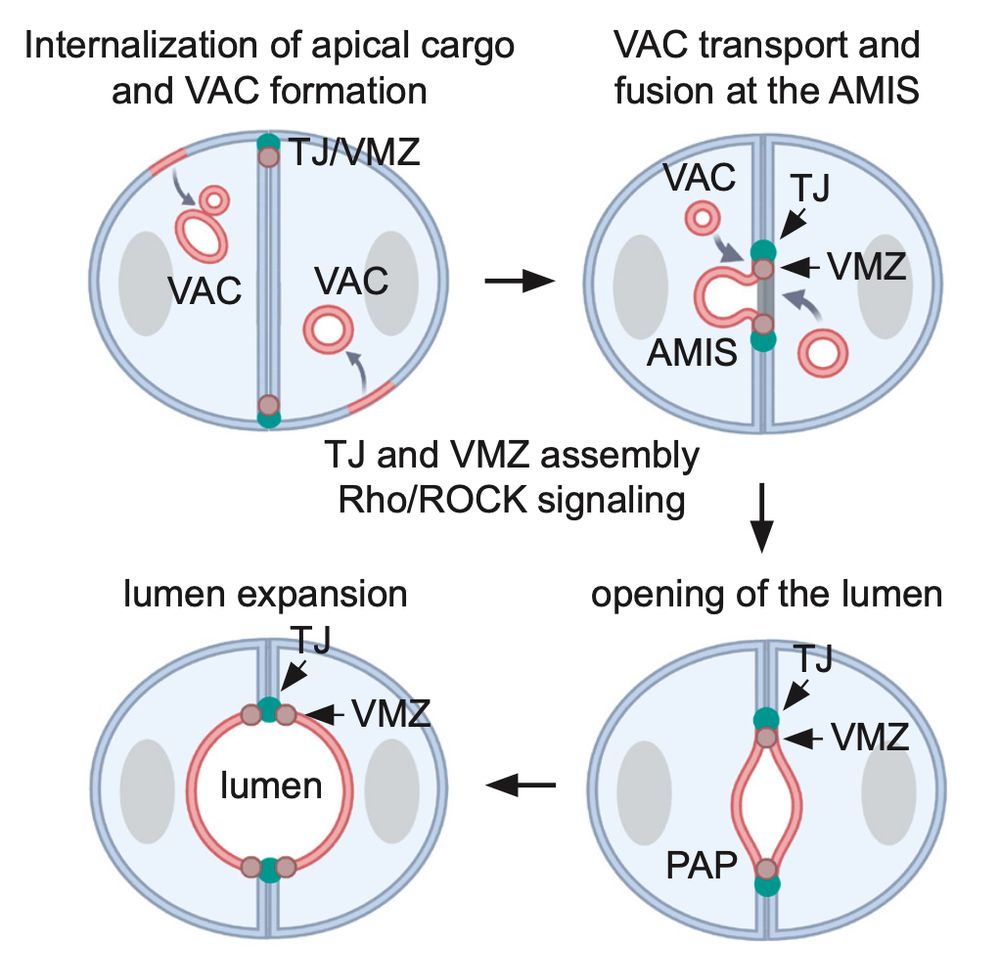
Altogether we propose that the fusion of VACs with the AMIS provides an efficient mechanism for the rapid specification of apical domain identity. In addition, Pals1 and PatJ appear to be critical for VAC fusion at the AMIS, possibly due to their role in regulating the apical actin cytoskeleton.
07.03.2025 07:40 — 👍 2 🔁 0 💬 0 📌 0
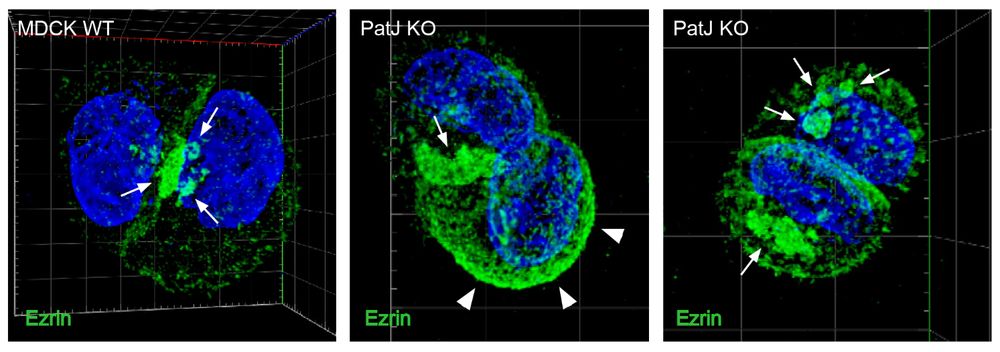
Finally, we asked how VAC exocytosis at the AMIS is regulated and found that loss of the Crb complex proteins Pals1 or PatJ results in the intracellular accumulation of VACs and severe defects in lumen formation. Apical actin levels and the apical cell cortex were markedly altered in PatJ KO cells.
07.03.2025 07:40 — 👍 1 🔁 0 💬 1 📌 0
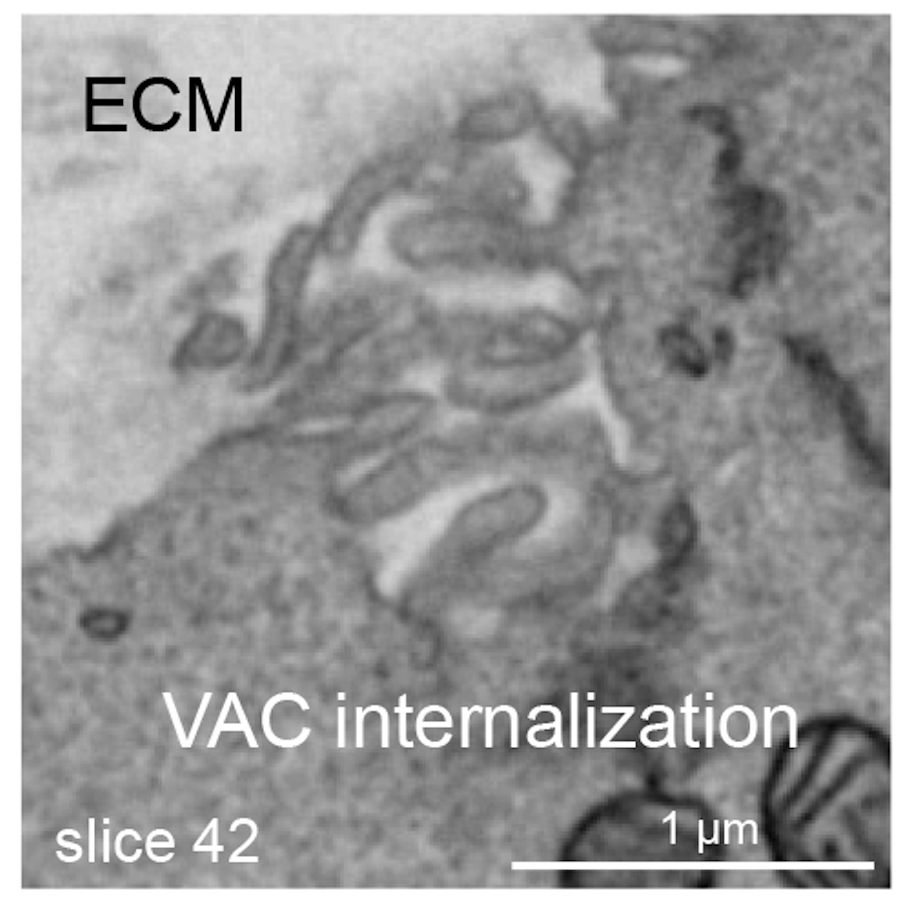
We further present evidence that VACs arise from the internalisation of microvilli-enriched membrane pits at the ECM facing side of cell doublets. How these membrane pits are internalised and how VACs are trafficked to and fuse with the AMIS is currently unclear.
07.03.2025 07:40 — 👍 0 🔁 0 💬 1 📌 0
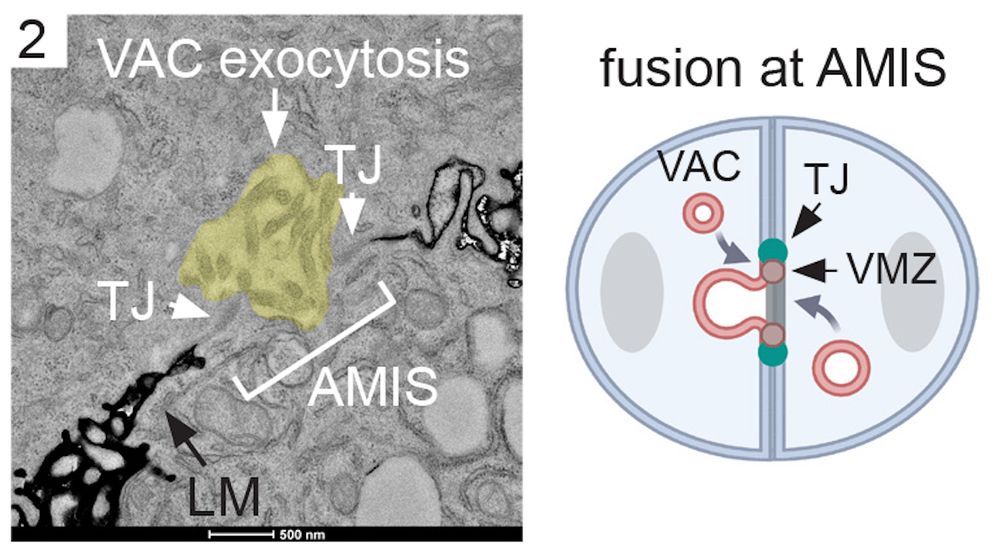
We then analysed lumen initiation in MDCK 3D cultures. Using LM and EM we identified intracellular VACs and observed fusion events of VACs with the Apical Membrane Initiation Site (AMIS). Interestingly, VAC fusion at the AMIS was again coordinated with the formation of cell-cell junctions.
07.03.2025 07:40 — 👍 0 🔁 0 💬 1 📌 0
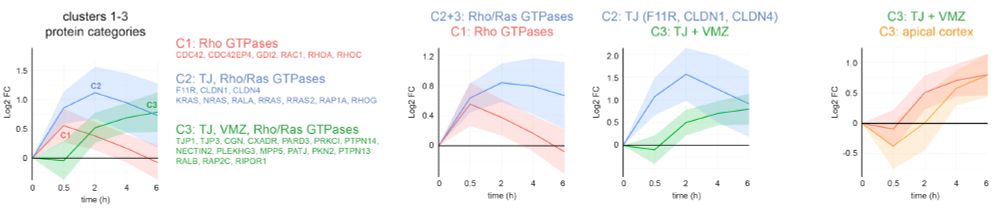
Interestingly, our time-resolved proteome revealed distinct recruitment profiles of Rho and Ras GTPases, tight junction membrane and scaffolding proteins, as well as apical proteins, providing new insight into the temporal execution of cell polarity development.
07.03.2025 07:40 — 👍 0 🔁 0 💬 1 📌 0
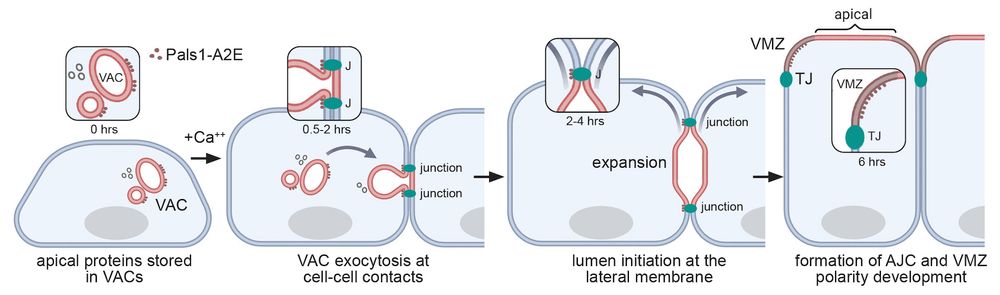
This set of experiments demonstrated that apical lumens are initiated by the exocytosis of large intracellular organelles at nascent cell-cell contact sites. Such organelles contain a pre-assembled microvilli-rich cortex and are known as Vacuolar Apical Compartments (VACs), or apicosomes.
07.03.2025 07:40 — 👍 0 🔁 0 💬 1 📌 0
To understand how apical lumen initiation and cell-cell junction assembly are coordinated, we initially analysed polarity development using the calcium switch assay, serial section TEM, and time-resolved proximity proteomics with our favourite enzyme APEX2. This is all done in MDCK cells.
07.03.2025 07:40 — 👍 0 🔁 0 💬 1 📌 0
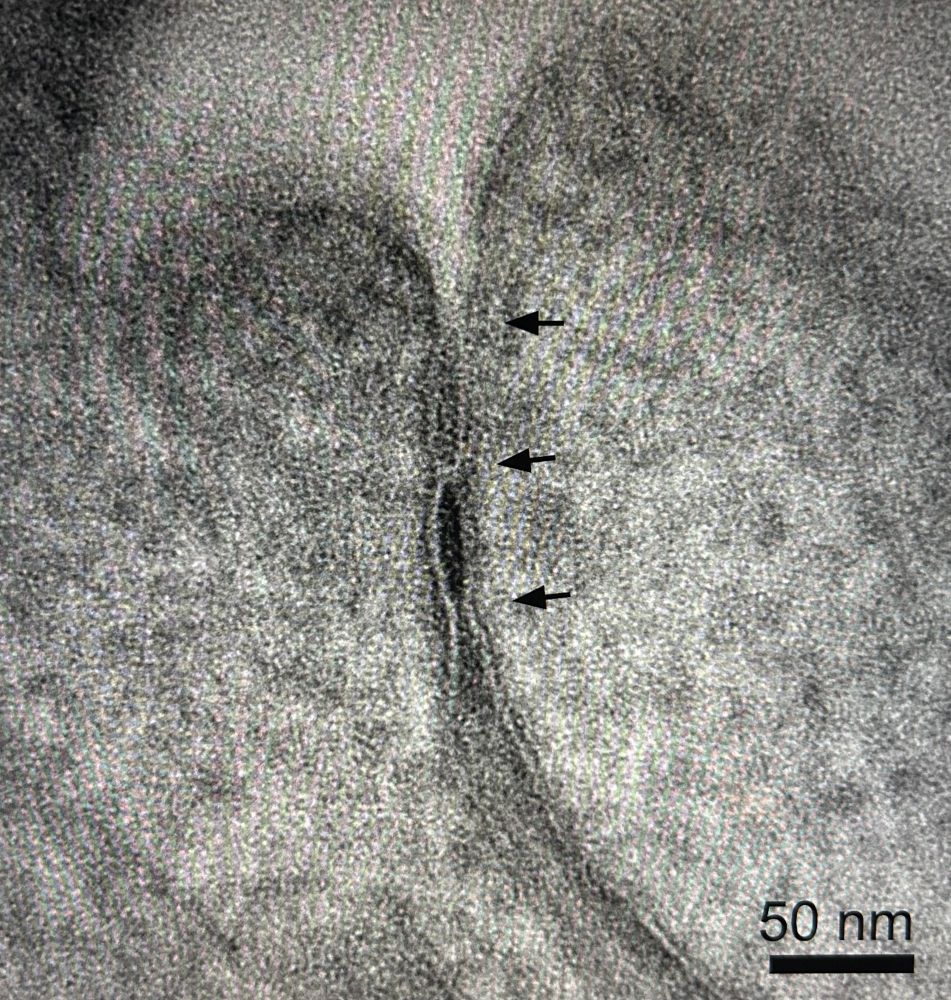
Another beautiful tight junction. This time with scale bar :)
06.02.2025 13:47 — 👍 8 🔁 1 💬 0 📌 0
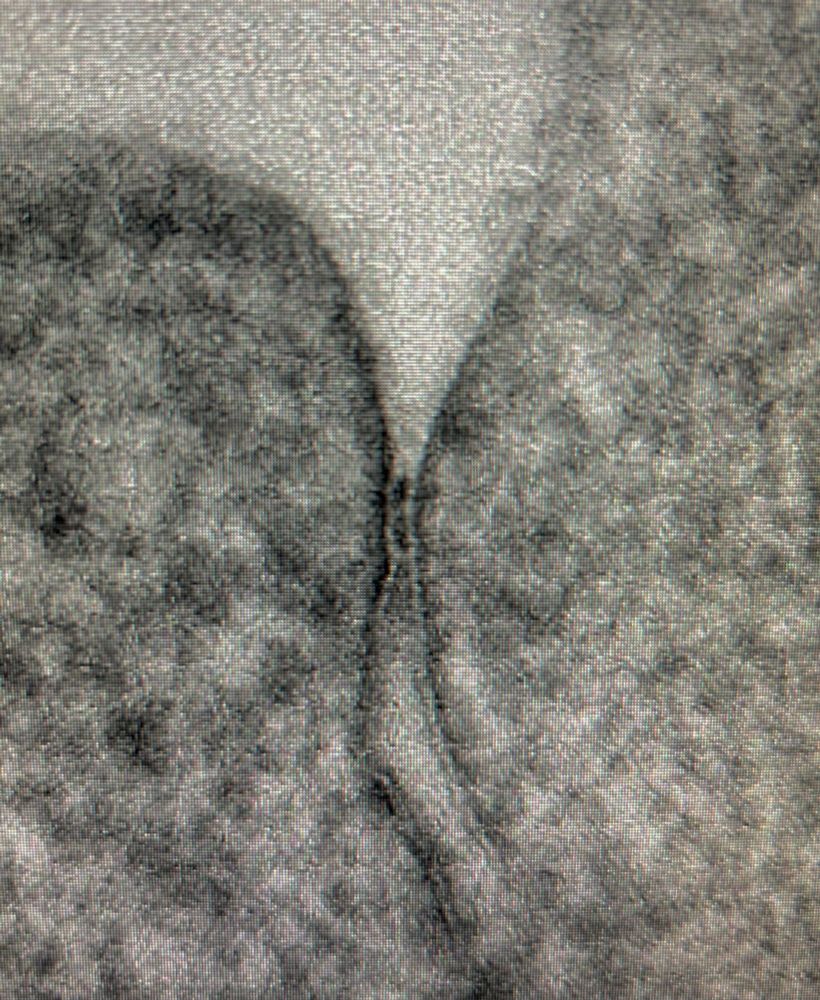
Not bad for a conventionally fixed and dehydrated TEM sample. No HPF/FS. OTO method. Stains the kissing points quite nicely.
02.02.2025 09:59 — 👍 13 🔁 0 💬 0 📌 0
First official post! A new paper out about #cytokinesis in #epithelia
www.embopress.org/doi/full/10....
Explore our journey as we uncover how regulators of cell-cell and cell-matrix interactions modulate cytokinesis efficiency, while revealing new roles for the Dystrophin-Dystroglycan complex.
15.11.2024 18:41 — 👍 49 🔁 14 💬 1 📌 0
This is sabotage! And this may well be just the beginning.
23.01.2025 00:31 — 👍 0 🔁 0 💬 0 📌 0
You don’t have to be German to know what this stands for - it’s clear as mud - and it’s disgusting!
21.01.2025 14:55 — 👍 1 🔁 0 💬 0 📌 0
You don’t have to be German to know what this means - it’s clear as mud - and it’s disgusting
21.01.2025 14:54 — 👍 2 🔁 0 💬 0 📌 0
iAPEX: Improved APEX-based proximity labeling for subcellular proteomics using an enzymatic reaction cascade https://www.biorxiv.org/content/10.1101/2025.01.10.632381v1
11.01.2025 08:46 — 👍 12 🔁 8 💬 0 📌 0
Our #CryoET paper on ciliary rootlet ultrastructure from mouse retina is now out in @elife.bsky.social! Have a peek with this video or dive deeper at bit.ly/3ZJzhgc. Big thanks to @carter-lab.bsky.social and #teamtomo.
#CryoET #CryoEM #Cilia #Rootlet #Centriole #Research #Science
08.12.2024 14:38 — 👍 100 🔁 34 💬 3 📌 3
Beautiful! Great work
03.12.2024 01:30 — 👍 1 🔁 0 💬 1 📌 0
Mind blowing! Look at those nuclear pores.
29.11.2024 00:10 — 👍 2 🔁 0 💬 0 📌 0
Would love to be added here - thank you!
20.11.2024 01:32 — 👍 1 🔁 0 💬 1 📌 0
Still one of my favourite tomograms, recorded ages ago at NCMIR (UCSD). It shows interconnected networks of caveolae (labeled with a miniSOG probe) at the rear of RPE1 cells. Bummer we so far haven't been able to image such networks in larger cell volumes; it's a lot of membrane that's stored here.
16.11.2024 03:42 — 👍 9 🔁 0 💬 0 📌 0
Oh wtf
15.11.2024 12:20 — 👍 1 🔁 0 💬 0 📌 0
oh... what happened here? why overlapping holes? two grids on top of each other?
15.11.2024 11:50 — 👍 1 🔁 0 💬 1 📌 0
Discovering the remarkable intracellular dynamics of killer T cells
Marie Curie Postdoc Fellow at Utrecht University 🇳🇱 Protein-Biochemist diving into cryo-ET ❄️
Studying collective cell behaviour.
Interested in regeneration, detection of aberrant cells and tissue mechanics.
(run by the Classenlab collective)
Biophysics / Cell Migration / Active matter / Mechanobiology
https://mpzpm.mpg.de/research/benoit-ladoux
https://ladoux-mege-lab.cnrs.fr
Postdoc at Göttingen CIDBN with Fred Wolf
Developmental Biology
Computational and Theoretical Biophysics
Statistical Inference
HetCCI is a DFG-funded Germany-wide research consortium (Priority Programme SPP 2493: Heterotypic cell-cell interactions in epithelial tissues). Website: https://www.uni-saarland.de/hetcci. Coordination and posts: Sandra Iden
Molecular logic of complex cell behaviors.
Orion Weiner's lab at UCSF. Account run by lab members.
#CellMigration #CellPolarity #Mechanics #OptoTools
weinerlab.com
Membrane cytoskeleton, specifically septins. Nanobodies, superresolution. Developing microscopy assays for cell biology. We love complicated experiments! Opinions my own as scientist, parent, citizen.
Since 1889 🗞️
Sign up for our newsletters and alerts: http://wsj.com/newsletters
Got a tip? http://wsj.com/tips
Follow our staff: https://go.bsky.app/2ppWqxF
Biophysics PhD Candidate at Mechanobiology Institute, National University of Singapore @ronglabofficial.bsky.social
Advancing cell biology in Singapore
The Society for Cell Biology Singapore (SCBS) was founded in 2022: “To promote the status and advance the interests of cell biology and its applications”.
www.scbs.sg
Asst Prof at UT Austin, Molecular Engineer, Tissue Aficionado, Former Drummer, belardi.che.utexas.edu
https://isb.med.upenn.edu/
Postdoc at UNC Chapel Hill • American Cancer Society, L’Oréal FWIS, & Leading Edge Fellow • Stony Brook University alum • Passionate about cell & developmental biology, C. elegans, science communication, & outreach • tmedwigkinney.com
🇪🇸 Postdoc researcher | ESPRIT Fellow @ BOKU University 🇦🇹
🔬 Plant cell biologist uncovering how plants defend their vacuoles and how their vacuoles defend them
🧬 Exploring non-canonical autophagy, ATG8ylation & Vacuolar Quality Control
Interdisciplinary research in Biophysics / Microscopy / Cell biology / Bioinformatics - lecturer in Biostatistics
IBDM - Aix-Marseille University
#Mechanics of #zebrafish, #ascidian, and #drosophila #development @Institute of Science and Technology Austria (ISTA)
https://heisenberglab.pages.ist.ac.at
Trends in Cell Biology is a leading reviews journal published by Cell Press covering the latest advances in cell biology. Editor Ilaria Carnevale, PhD.
cryoEM, structural biology, ribosomes
https://www.genzentrum.uni-muenchen.de/research-groups/beckmann/index.html