Only in this collision geometry do the positioning of the two RACK1s allow ZAK’s RACK1-interacting motif (RIM) to engage them and enabling its SAM domains to dimerize — the key step for activation. This activation is further negatively regulated by SERBP1 competing for the RACK1 interface. 4/4
23.11.2025 15:52 —
👍 3
🔁 0
💬 0
📌 0
ZAK is constitutively recruited to ribosomes via its RACK1-interacting helix (RIH), the eS27-pin, and the ES7-contacting patch. Upon stress, ribosome collisions generate a unique interface that enables specific contacts — the ES6-patch and the RIH-peptide. 3/4
23.11.2025 15:47 —
👍 3
🔁 0
💬 1
📌 0
Despite being the driving force behind the RSR, the interaction between ZAK and ribosomes has remained elusive for years. Here, we show how ZAK is recruited to ribosomes in native as well as stress-induced conditions, and how collision-specific interactions organized on RACK1 mediate activation. 2/4
23.11.2025 15:47 —
👍 2
🔁 0
💬 1
📌 0
How does the kinase ZAK sense ribosome collisions? Find out in our latest collaboration with the @greenlab.bsky.social @doubleshuang.bsky.social @Vienna Huso: 1/4
rdcu.be/eRmJl
#ribosome #cryoEM #LMU #JHMI
23.11.2025 15:46 —
👍 49
🔁 20
💬 2
📌 2
Despite being the driving force behind the RSR, the interaction between ZAK and ribosomes has remained elusive for years. Here, we show how ZAK is recruited to ribosomes in native as well as stress-induced conditions, and how collision-specific interactions organized on RACK1 mediate activation. 2/4
23.11.2025 14:06 —
👍 3
🔁 0
💬 0
📌 0
Over the next six years, we’ll uncover the role of snoRNPs in ribosome assembly and disease. This project will be led in our lab by Roland Beckmann and Matthias Thoms @thomsmatt.bsky.social #ribosome #cryoEM
07.11.2025 13:50 —
👍 6
🔁 0
💬 0
📌 0

🎉Thrilled to share that our lab has been awarded an ERC Synergy Grant @erc.europa.eu! Excited for amazing collaborations with the Pertschy (Uni Graz), Henras (CBI Toulouse), and Woodson (Johns Hopkins) labs.
#ERCSyG #GeneCenter #LMU #UniGraz #CNRS #JohnsHopkins
07.11.2025 13:50 —
👍 26
🔁 5
💬 4
📌 0
We are excited to share our collab with @kedrov-lab.bsky.social! Using Cryo-EM❄️ on an amazing sample, we found that YidC is recruited to the back of SecYEG during late-stage substrate insertion —supporting that the "back-of-Sec" route observed in eukaryotes also exists in bacteria 🦠. Check it out👇👇👇
28.05.2025 08:22 —
👍 20
🔁 4
💬 0
📌 1

Substrate-induced assembly and functional mechanism of the bacterial membrane protein insertase SecYEG-YidC
The universally conserved Sec translocon and the YidC/Oxa1-type insertases mediate biogenesis of alpha-helical membrane proteins, but the molecular basis of their cooperation has remained disputed over decades. A recent discovery of a multi-subunit insertase in eukaryotes has raised the question about the architecture of the putative bacterial ortholog SecYEG-YidC and its functional mechanism. Here, we combine cryogenic electron microscopy with cell-free protein synthesis in nanodiscs to visualize biogenesis of the polytopic membrane protein NuoK, the subunit K of NADH-quinone oxidoreductase, that requires both SecYEG and YidC for insertion. We demonstrate that YidC is recruited to the back of the translocon at the late stage of the substrate insertion, in resemblance to the eukaryotic system, and in vivo experiments indicate that the complex assembly is vital for the cells. The nascent chain does not utilize the lateral gate of SecYEG, but enters the lipid membrane at the SecYE-YidC interface, with YidC being the primary insertase. SecYEG-YidC complex promotes folding of the nascent helices at the interface prior their insertion, so the examined cellular pathway follows the fundamental thermodynamic principles of membrane protein folding. Our data provide the first detailed insight on the elusive insertase machinery in the physiologically relevant environment, highlight the importance of the nascent chain for its assembly, and prove the evolutionary conservation of the gate-independent insertion route. ### Competing Interest Statement The authors have declared no competing interest. Deutsche Forschungsgemeinschaft, https://ror.org/018mejw64, Ke1879/3, 267205415 (CRC 1208) European Research Council, https://ror.org/0472cxd90, CRYOTRANSLATION
Very special feelings to announce this one... A project that started like 10 years ago is reaching the finish line, ready to shine. In a dream-team with @beckmannlab.bsky.social we solved the long-chased structure of the active membrane protein insertase SecYEG-YidC
www.biorxiv.org/content/10.1...
27.05.2025 09:21 —
👍 60
🔁 24
💬 7
📌 7
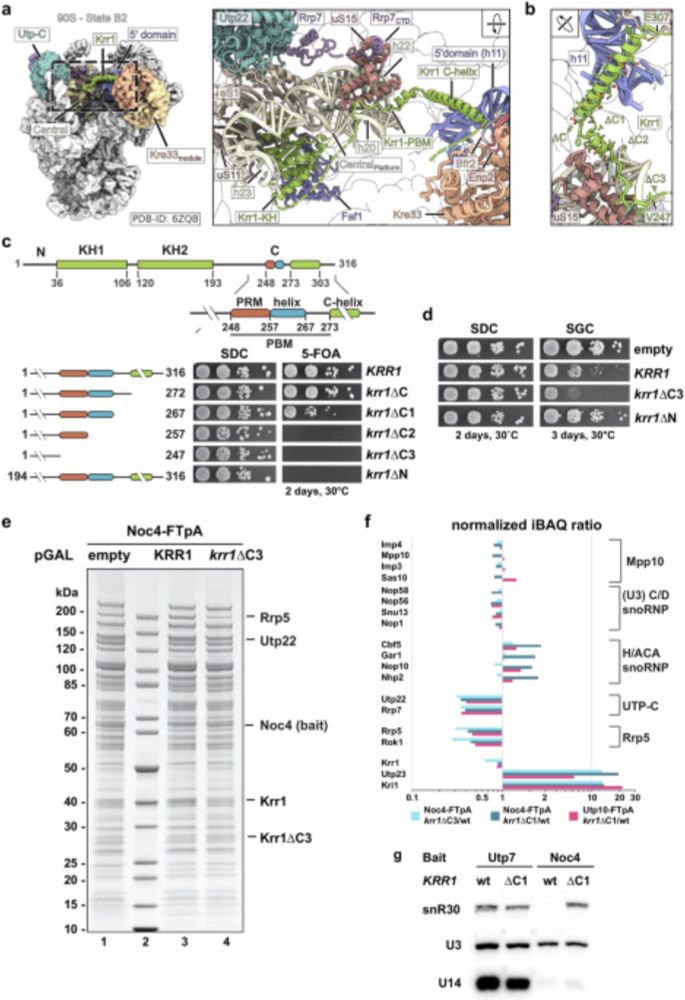
We are happy to share that our snR30 story is finally out in @natureportfolio.nature.com 🥳 We report the first structure of a H/ACA snoRNP acting in ribosome synthesis thereby providing a detailed structural and biochemical view of the snR30 snoRNP guiding local 18S rRNA subdomain folding. 👇👇👇
26.05.2025 09:29 —
👍 62
🔁 27
💬 1
📌 3
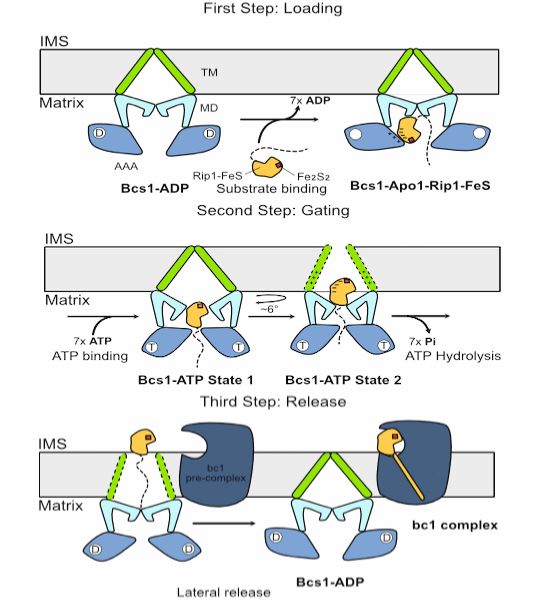
How does an unusual heptameric AAA-ATPase translocate fully-folded Rieske protein across the inner mitochondrial membrane?
Cryo-EM & functional data by Roland Beckmann and coworkers show how Bcs1 alternates between different states during its translocation activity.
www.embopress.org/doi/full/10....
23.05.2025 11:23 —
👍 16
🔁 8
💬 4
📌 1