
A prophage-encoded abortive infection protein preserves host and prophage spread www.nature.com/articles/s41...
28.01.2026 19:13 — 👍 42 🔁 20 💬 0 📌 2@owentuck.bsky.social
chem bio PhD student in the Doudna Lab @Berkeley

A prophage-encoded abortive infection protein preserves host and prophage spread www.nature.com/articles/s41...
28.01.2026 19:13 — 👍 42 🔁 20 💬 0 📌 2
NLR-like immunity in bacteria
A new study from the Alex Gao lab. The scope of this work is incredible!!!
www.biorxiv.org/content/10.6...

Bacterial genomes encode a rich repertoire of antiphage systems, but we still know surprisingly little about when these systems are actually expressed.
In this preprint, Lucas Paoli et al, ask what shapes antiphage systems expression in native contexts.
www.biorxiv.org/content/10.6...

#microsky #phagesky
More anti #phage systems in E. coli
www.biorxiv.org/content/10.6...

A very nice Preview of our work in
@cp-cellhostmicrobe.bsky.social
this morning from Chrishan Fernando & Nicole D. Marino! www.cell.com/cell-host-mi...
✨New preprint!
🧵1/4 Excited to share our work on AI-guided design of minimal RNA-guided nucleases. Amazing work by @petrskopintsev.bsky.social @isabelesain.bsky.social @evandeturk.bsky.social et al!
Multi-lab collaboration @banfieldlab.bsky.social @jhdcate.bsky.social @jacobsenucla.bsky.social🧬
🔗👇

I’m happy to share our new preprint! We uncovered the full diversity of bacterial TIR-based antiviral immune signaling, massively expanded the known diversity of Thoeris systems, and revealed conservation of TIR-derived immune signals across the tree of life.
www.biorxiv.org/content/10.6...

🚨Preprint alert - this is a big one! We transfer the revolutionary power of TnSeq to bacteriophages.
Our HIDEN-SEQ links the "dark matter" genes of your favorite phage to any selectable phenotype, guiding the path from fun observations to molecular mechanisms.
A thread 1/8

I am so excited to share our project with you! We find prokaryotic proteases activate toxic enzymes and pores as a modular strategy in phage defense. We studied four fascinating protease-toxin pairs that are abundant across bacterial genomes:
www.biorxiv.org/content/10.1...
Beautiful preprint from Simone Evans et al. in Alex Gao's group looking at MBL/nuclease and other cool zymogens (pepco, EACC1) in antiphage defense systems. Great to see this paradigm extended - probably many more proteolytically activated effectors out there...
www.biorxiv.org/content/10.1...
Thank you Aude!!
15.11.2025 21:51 — 👍 0 🔁 0 💬 0 📌 0Thanks Cress friends!
14.11.2025 18:09 — 👍 1 🔁 0 💬 0 📌 0Thanks Jason! We miss you
14.11.2025 17:46 — 👍 1 🔁 0 💬 0 📌 0Fascinating discovery - anti-phage defense protein is a proenzyme that is cleaved by partner protease after phage infection and all three! products of cleavage form active nuclease.
14.11.2025 07:51 — 👍 9 🔁 5 💬 0 📌 0Thanks Luuk!
14.11.2025 17:44 — 👍 0 🔁 0 💬 0 📌 0Thanks François - your work was influential in our thinking on this project (and others)
14.11.2025 17:43 — 👍 3 🔁 0 💬 0 📌 0
Our nuclease-protease story is out! We explored a fascinating case of coevolution and modularity in prokaryotic immune systems: www.science.org/doi/10.1126/...
Thanks to wonderful coauthors/collaborators/friends, the whole @doudna-lab.bsky.social and everyone at @innovativegenomics.bsky.social

Very excited to share the latest work from our lab, which was published today in Nature!
nature.com/articles/s41...
PhD graduate and now post-doc Sofia Dahlman, along with co-senior author Sam Forster from The Hudson and other researchers from our lab and others.

New pre-print from the Banfield lab, highlighting an interesting case of 1.5Mb megaplasmids found in human gut.
Plasmid genomes were resolved using #PacBio HiFi sequencing with hifiasm-meta for #metagenome assembly. Host association was detected using epigenetic signals.
doi.org/10.1101/2025...

Today in @nature.com , we highlight how a cousin of CRISPR-Cas10, mCpol, establishes an evolutionary trap in anti-phage immune systems.
Check out @erinedoherty.bsky.social and my work from @doudna-lab.bsky.social lab here:
www.nature.com/articles/s41...

A recent cool preprint by John Whitney's lab on a new family of antibacterial proteins secreted by Gram-positive bacteria that enter and kill a broad spectrum of bacteria. Cell entry is receptor-independent and relies on cleavage by a co-secreted protease and the PMF.
www.biorxiv.org/content/10.1...


Some pics of our awesome PhD candidate, @sophswartz.bsky.social, presenting at the Cold Spring Harbor Laboratory conference!
25.08.2025 20:43 — 👍 10 🔁 1 💬 0 📌 0
Excited to share our new preprint co-led by @jnoms.bsky.social!
Here we reveal an exceptional diversity of viral 2H phosphodiesterases (PDEs) that enable immune evasion by selectively degrading oligonucleotide-based messengers. This 2H PDE fold has evolved striking substrate breath & specificity.
Congrats and best of luck!
18.08.2025 19:54 — 👍 2 🔁 0 💬 1 📌 0Very happy to share that I will be starting my lab at AITHYRA in October! My lab will use structural bioinformatics and functional genomics to understand the function of viral proteins, with a special emphasis on understanding how viruses subvert innate immunity.
15.08.2025 19:54 — 👍 36 🔁 5 💬 4 📌 1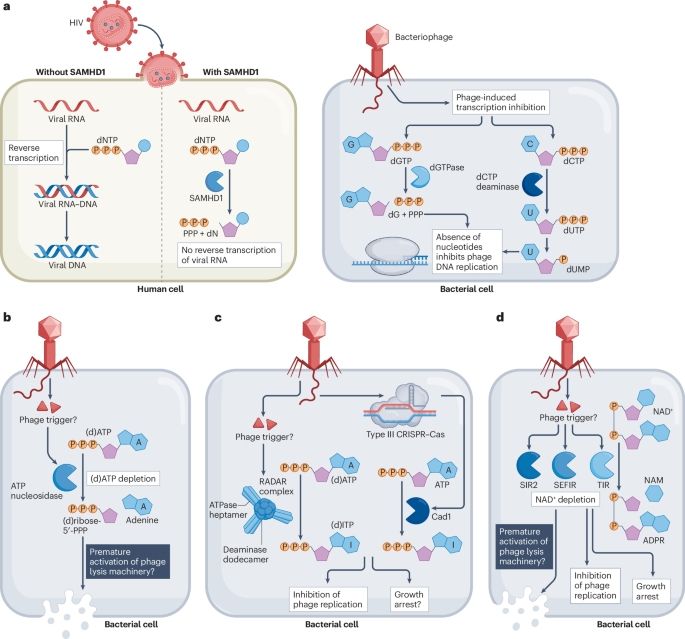
We wrote a review on the free nucleotide pool as a central playground in human, bacterial, and plant immunity – now out in Nature Reviews in Immunology
www.nature.com/articles/s41...
Was fun to write this piece with Dina Hochhauser!
Here is a thread to explain the premises
1/
‼️ New pre-print from co-leads @owentuck.bsky.social and Jason Hu! Check out this fascinating example of how coevolution enables defense system innovation.
29.07.2025 21:18 — 👍 26 🔁 9 💬 0 📌 0Thanks Luuk!
29.07.2025 19:06 — 👍 1 🔁 0 💬 0 📌 0Absolutely! Caspases seem particularly useful for proenzyme activation… 🧐
29.07.2025 18:44 — 👍 1 🔁 0 💬 0 📌 0Congratulations!
29.07.2025 18:43 — 👍 1 🔁 0 💬 0 📌 0