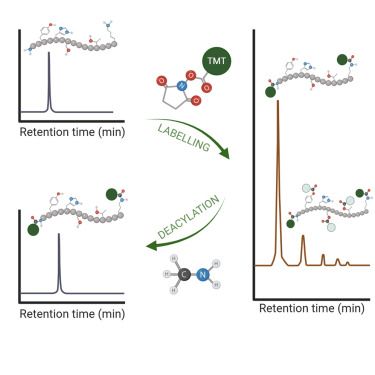
Looking forward to an exciting time at @hupo-org.bsky.social world congress in Toronto! swing by my poster on Monday to know more about dynamic proteomic screen of E3 ligases. #HUPO2025 #teamMassSpec #proteomics
07.11.2025 07:53 — 👍 0 🔁 0 💬 0 📌 0

mzPeak: Designing a Scalable, Interoperable, and Future-Ready Mass Spectrometry Data Format
Advances in mass spectrometry (MS) instrumentation, including higher resolution, faster scan speeds, and improved sensitivity, have dramatically increased the data volume and complexity. The adoption of imaging and ion mobility further amplifies these challenges in proteomics, metabolomics, and lipidomics. Current open formats such as mzML and imzML struggle to keep pace due to large file sizes, slow data access, and limited metadata support. Vendor-specific formats offer faster access but lack interoperability and long-term archival guarantees. We here lay the groundwork for mzPeak, a next-generation community data format designed to address these challenges and support high-throughput, multidimensional MS workflows. By adopting a hybrid model that combines efficient binary storage for numerical data and both human- and machine-readable metadata storage, mzPeak will reduce file sizes, accelerate data access, and offer a scalable, adaptable solution for evolving MS technologies. For researchers, mzPeak will support complex workflows and regulatory compliance through faster access, improved metadata, and interoperability. For vendors, it offers a streamlined, open alternative to proprietary formats. mzPeak aims to become a cornerstone of MS data management, enabling sustainable, high-performance solutions for future data types and fostering collaboration across the mass spectrometry community.
I'm a huge fan of this proposal!
If we can get vendor buy-in to an open, common format, it could bring more computational folks into the field. How can these folks be excited about #proteomics when its hard to even read the already complicated data?
pubs.acs.org/doi/10.1021/...
10.10.2025 17:53 — 👍 21 🔁 4 💬 3 📌 2

Postdoctoral Research Fellow (f/m/d)
Postdoc: Investigate the role of translational regulation in controlling cell resilience,sequencing- and mass spectrometry,independently analyze the resulting data
My lab is looking for an enthusiastic postdoc who is motivated to grow with our team and passionate about research on cellular resilience. If you are interested, or know someone who might be, please apply through the link below.
06.10.2025 12:59 — 👍 1 🔁 1 💬 0 📌 0

Gearing up for what promises to be an exciting week ahead @crg.eu #CRGtraining #teamMassSpec
06.10.2025 07:38 — 👍 14 🔁 1 💬 0 📌 0
The location and food will make it impossible for anyone else to ever make a better EUPA conference. Thank you for organising such an excellent event! #EUPA2025
19.06.2025 11:24 — 👍 8 🔁 1 💬 1 📌 0
Thank you all the colleagues that reached out in person and via DMs to share their thoughts, organisers for giving me the opportunity and amazing chair Martha Ingola and @mtrost.bsky.social
18.06.2025 11:47 — 👍 1 🔁 0 💬 0 📌 0

How often do you get to talk at a conference with views like this? Had an amazing time sharing my research at #eupa2025.
18.06.2025 11:45 — 👍 0 🔁 0 💬 1 📌 0

Gearing up for #EuPA2025 next week & super excited to share my work on combining siRNA screen with translation/degradation proteomics. I mean, look at that auditorium!! 😮 Want to know how you can do high-throughput proteomics without a liquid handling robot or a high speed TOF like its 2020? join me
10.06.2025 12:52 — 👍 1 🔁 0 💬 0 📌 0
Very excited for Fast & Fair peer review - authors get decisions, with reviews, within 7 business days of submission. Reviews are high quality - lots of quality control by our academic editors, plus we pay reviewers.
25.03.2025 11:25 — 👍 29 🔁 9 💬 0 📌 1
🤷♂️ the company rep says you can reuse them and we’ve had a good experience working with 100ng-35ug. But perhaps making your own tips might be cheaper.
24.03.2025 14:16 — 👍 1 🔁 0 💬 0 📌 0
Affinisep
24.03.2025 07:34 — 👍 1 🔁 0 💬 1 📌 0
During Masters thesis under Prof. Paul Haynes and Prof. Mark Molloy (Macquarie University) circa 2012-14, and a lot more as a technician at a core facility 2014-17 with @itsbinir.bsky.social hanging around.
20.03.2025 13:35 — 👍 3 🔁 0 💬 0 📌 0
Not sure, but we usually get >90% 0 missed cleavage with STrap and SP3 at 47C, 60 mins in 50mM TEAB/EPPS.
18.03.2025 18:51 — 👍 2 🔁 0 💬 0 📌 0
The analogy of omics data normalization and cooking:
some processing is usually necessary, overcooking makes it bland, highly over-processed foods are unhealthy, and the quality of the ingredients matters.
27.02.2025 08:01 — 👍 76 🔁 23 💬 1 📌 5
What is up with @novonordisk.bsky.social ? 📉
15.02.2025 15:18 — 👍 0 🔁 0 💬 0 📌 0
Congrats Tina!
10.02.2025 19:24 — 👍 0 🔁 0 💬 0 📌 0
Trying to find who is, and how. But I’m afraid many tribrids and QEs will need to be retired and replaced with TOFs if labs/core facilities are to keep up.
08.01.2025 00:10 — 👍 0 🔁 0 💬 0 📌 0
Any specific reason for OT-OT instead of OT-IT?
07.01.2025 17:34 — 👍 0 🔁 0 💬 1 📌 0
Haven’t tried DIA on lumos but I’m inclined to believe 2hrs/sample with FAIMS-OT-IT might actually be faster for the same quant ids.
07.01.2025 14:33 — 👍 0 🔁 0 💬 1 📌 0
Similar for us on ascend but for lumos which doesn’t support RTS, SPS-MS3 with 24 TMT18 fractions ends up being 2 days for about 8k+ quant. With large batches and IRS channel, the throughput feels very 2018(?).
07.01.2025 14:33 — 👍 1 🔁 0 💬 1 📌 0
‘30spd’ == anyone getting TMT16 or TMT32 fractions finished within a day with >8k proteins?
06.01.2025 17:54 — 👍 0 🔁 0 💬 1 📌 0
Hey #TeamMassSpec, with all the super duper ultra fast TOF based DIA methods out there, how are you managing the throughput & depth on your old tribrids, QEs and Exploris? Any TMT aficionados out here getting 30spd, for instance, or even 15 with >8k proteins?
06.01.2025 17:50 — 👍 3 🔁 4 💬 3 📌 0
Happy holidays, and a reminder to ALWAYS check your synthesised peptide standards for isotope purity. #teammassspec
24.12.2024 21:48 — 👍 2 🔁 0 💬 0 📌 0
#TeamMassSpec represent at the first symposium on advances in therapeutic approaches.
09.12.2024 07:16 — 👍 0 🔁 0 💬 0 📌 0

Forgot to introduce myself on here so here you go. Hello #teammassspec, I’m a 3rd year postdoc with 13 years of experience doing proteomics at all career stages (MSc, technical scientist, JRF, PhD). India>Australia>Belgium>Germany. Here’s me a couple of weeks ago at #HUPO Dresden
17.11.2024 16:16 — 👍 4 🔁 0 💬 0 📌 0
Hi Alejandro, would love to be added too. Cheers!
17.11.2024 16:02 — 👍 1 🔁 0 💬 0 📌 0
Postdoc @ Küster lab | prev, PhD @ Trost lab | proteomics | chemoproteomics | mass spectrometry | target deconvolution | drug MoA
Associate Professor at UCSF, visiting scientist at the Gladstone Institutes. Expertise in mass spectrometry and proteomics. Views are my own. profiles.ucsf.edu/danielle.swaney
Mass spectrometry for high-throughput quantitative proteomics with special interest in posttranslational modifications.
Proteomics at Spanish National Centre for Cardiovascular Research
Chief Science and Strategy Officer, openRxiv. Co-Founder, bioRxiv and medRxiv.
...we are passionate about spatial proteomics, mass spectrometry and single-cell technologies to study disease mechanisms @MDC_Berlin 🇩🇪🇨🇴🇨🇳🇰🇿🇵🇷🇵🇱🇲🇽
Old person forgetting everything they ever knew about proteomics at speeds in excess of 270 Hz...
The Human Proteome Organization (HUPO) is an international scientific organization representing and promoting proteomics through international cooperation and collaborations
Senior Scientist at Evosep, focus on developing workflows for mass spectrometry based biolomecule characterization and proteome profiling. 2 wheels + an engine = fun!
Protein biochemist | CNRS Researcher @IGBMC, Strasbourg, France | Quantitative and comparative interactomics, protein-protein interactions, domain-motif interactions, proteomics, variant effects
Proteomics, precision health, mass spectrometry, biomarkers, quantitative, changing medicine
Connecting the global TPD and ubiquitin biology community. Discover research labs, explore conferences, and stay in the loop on everything from E3 ligases to molecular glues.
TPD research map:
https://tpd-community.com/
Dietrich Volmer · Research in analytical chemistry, mass spec and metabolomics · Editor-in-Chief of Anal. Sci. Adv. & Rapid Commun. Mass Spectrom · Views are my own
Humboldt University · Berlin · 🇨🇦🇩🇪 · volmerlab.de
Research and development company leveraging the biology of diverse organisms. Read about our science: http://research.arcadiascience.com
Biologist, Professor at CECAD, University of Cologne.
We study aging, age-related diseases, stem cell biology and proteostasis.
http://www.davidvilchezlab.com/
Mass spec sitter at MRC LMB
Focus on developing proteomics technologies.
https://www.chem.ox.ac.uk/people/shabaz-mohammed
https://www.bioch.ox.ac.uk/research/mohammed
https://scholar.google.co.uk/citations?hl=en&user=6FdXeiwAAAAJ&view_op=list_works&sortby=pubdate
Research groups at @scilifelab.se in Stockholm and @mdc-berlin.bsky.social in Berlin. Doing biochemistry with #proteomics and #massspectrometry.
We seek principles in the coordination among protein synthesis, metabolism, cell growth and differentiation. PI: @slavov-n.bsky.social
Web: https://slavovlab.net
Videos: http://youtube.slavovlab.net