
Using CG methylation (mCG) of heterochromatic transposons as a trait for GWAS, we identified a major genetic association to an uncharacterized gene: a plant ortholog of vertebrate CDCA7.
07.11.2025 17:01 — 👍 6 🔁 2 💬 1 📌 0@jlawlab.bsky.social

Using CG methylation (mCG) of heterochromatic transposons as a trait for GWAS, we identified a major genetic association to an uncharacterized gene: a plant ortholog of vertebrate CDCA7.
07.11.2025 17:01 — 👍 6 🔁 2 💬 1 📌 0
And this great Nature Plants News & Views by Pandesha and Slotkin covering this breakthrough discovery
www.nature.com/articles/s41...
@hadse.bsky.social @KeithSlotkin @natplants.nature.com
8/8

Interested in learning even more?
Check out complementary work from Wu and Jacobsen revealing roles for a TF binding protein in this new mode of DNA methylation targeting. www.nature.com/articles/s41...
@jacobsenucla.bsky.social @ZhongshouWu
7/8

This new targeting mode for DNA methylation also opens new avenues for pursuing precise epigenomic editing to correcting DNA methylation defects and or engineer new epialleles for crop improvement.
6/8

This discovery transforms our view of how DNA methylation patterns are regulated in plants—moving from purely chromatin-driven models to an integrated framework where TFs guide methylation reprogramming during development.
5/8

This new TF-based mode of targeting controls the vast majority of siRNAs used by the RNA-directed DNA methylation (RdDM) pathway to target methylation in anthers and ovules, revealing it plays a massive role in shaping the epigenome in plant reproductive tissues.
4/8

Leveraging genome engineering, we demonstrated that the DNA motif bound by a RIM TF is required to target methylation and that mis-expression of this TF can remodel the epigenome in a locus- and tissue-specific manner. In short, the DNA motifs and TFs are necessary AND sufficient.
3/8

Using forward and reverse genetics approaches we identified several REM family TFs that Instruct DNA methylation, designated as RIMs. We showed that different RIMs regulate methylation patterns in male and female reproductive tissues.
2/8

Have you ever wondered how new DNA methylation patterns are established?
Paradigm shift ahead! We discovered a new mode of DNA methylation targeting in plants that relies on transcription factors and sequence motifs rather than chromatin modifications to regulate the methylome. rdcu.be/eQ6L5
1/8

Salk researchers are using a little flowering plant to answer the question: How do cells generate new epigenetic change? The answer marks a major shift in plant biology and may inform future epigenetic engineering strategies with medical and agricultural applications.
www.salk.edu/news-release...
Excited about the future of plant epigenetics?
Join us at the Keystone Symposium Plant Epigenetics & Epigenome Engineering in Fort Collins, October 13-16, 2025! keystonesymposia-19559593.hs-sites.com/%F0%9F%8C%BF...
Registration is open!
#PlantScience #Epigenetics #KeystoneSymposia
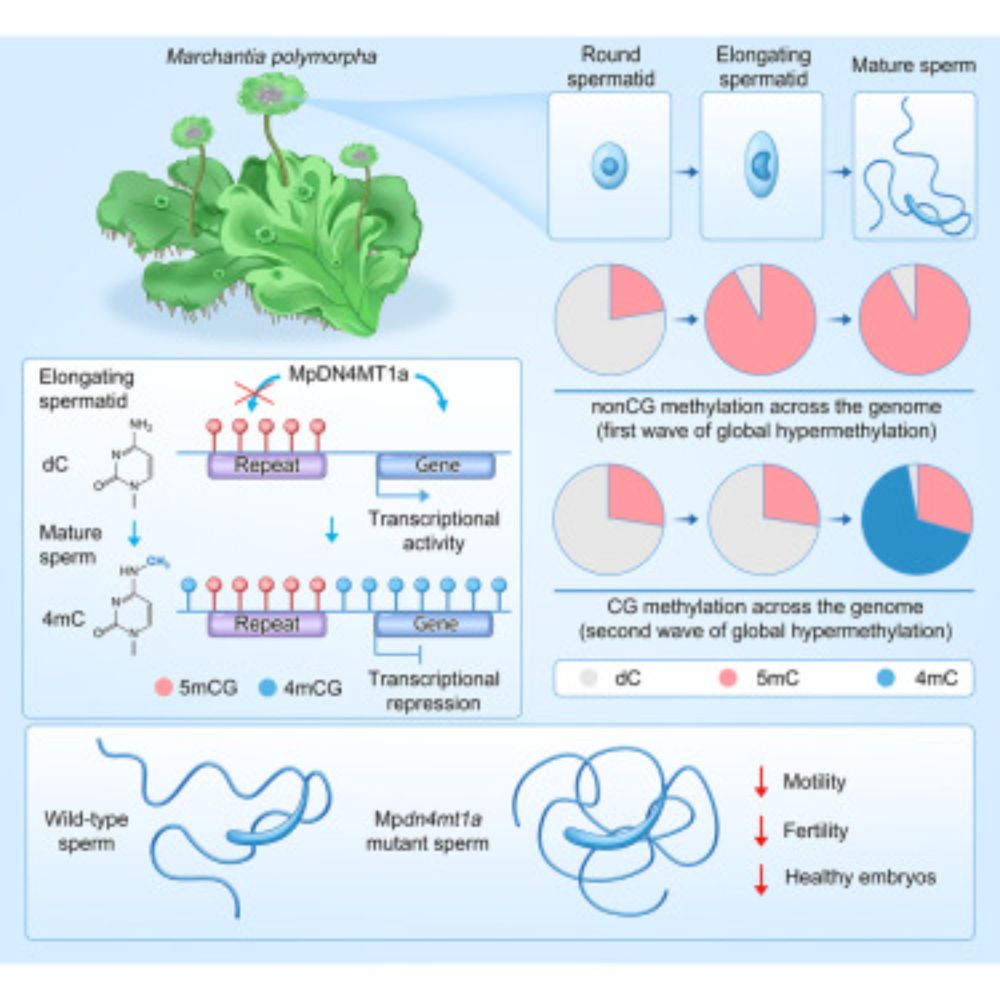
I'm thrilled to share that our study is now published in Cell:
Extensive N4 cytosine methylation is essential for Marchantia sperm function.
www.cell.com/cell/fulltex...
This paper confirms our 4mC discovery in Marchantia sperm and takes it much further.
A thread: 0/13

Excellent to see this amazing work out from James Walker and Xiaoqi Feng - amazing stuff!
Extensive N4 cytosine methylation is essential for Marchantia sperm function
www.cell.com/cell/fulltex...