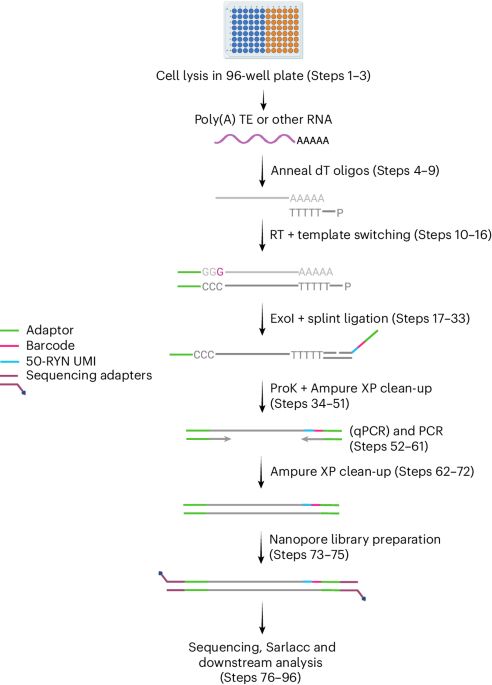
Have you ever wondered how new DNA methylation patterns are established?
Paradigm shift ahead! We discovered a new mode of DNA methylation targeting in plants that relies on transcription factors and sequence motifs rather than chromatin modifications to regulate the methylome. rdcu.be/eQ6L5
1/8
11.12.2025 21:56 — 👍 47 🔁 23 💬 2 📌 0
Excellent stuff @p-bourguet.bsky.social !
08.11.2025 20:57 — 👍 2 🔁 0 💬 1 📌 0

Excited to see our story on epigenetic mediated aclimation of plants to fluctuating light patterns out today in doi.org/10.1111/nph..... This work started 6 years ago co-supervised with the amazing @proftlawson.bsky.social and led by the brilliant @robynemm.bsky.social. 1/2
20.09.2025 12:04 — 👍 34 🔁 6 💬 2 📌 1
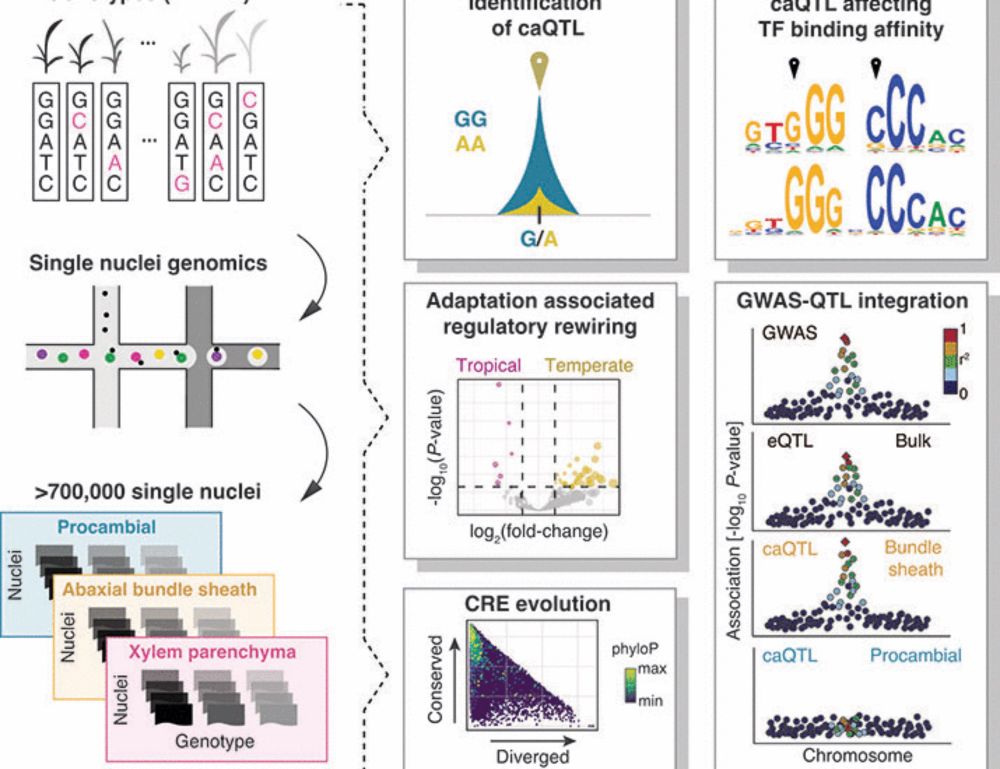
Dr Tina Schreier awarded ERC Starting Grant
@tinaschreier.bsky.social has been awarded an @erc.europa.eu Starting Grant for research into the cell biology of C4 photosynthesis 🌱
Tina’s project will explore the cell biology that leads to the formation of specialised bundle sheath cells in C4 plants 👇
bit.ly/4mWFGhh
04.09.2025 11:00 — 👍 17 🔁 3 💬 1 📌 0

We are pleased to share that P4S Research Fellow @jamespblloyd.bsky.social from the University of Western Australia was recently awarded a Grains Research and Development Corporation Mid-Career Research Fellowship 🌟
Read more 👉 www.linkedin.com/feed/update/...
29.07.2025 01:42 — 👍 13 🔁 3 💬 0 📌 0
#PlantBio2025
I’ll be speaking in the final plenary—sharing the story behind this paper and some new, unpublished work from my lab at TSL. Hope people stick around till the end (though a few already warned me they’re leaving early 😅)!
26.07.2025 17:09 — 👍 47 🔁 9 💬 2 📌 0

Research Fellow (Postdoctoral) at University of Birmingham
An opportunity for an academic position as a Research Fellow (Postdoctoral) is available, as advertised on jobs.ac.uk. Apply now and explore other academic job openings.
I have a Postdoctoral Research Fellow position starting in January to study the evolution of seed gene networks using the fern Ceratopteris! Interested? More info here:
www.jobs.ac.uk/job/DNU179/r...
Closing date 31st July. 🙂
#PlantScience
#PlantSciencejobs
10.07.2025 17:07 — 👍 27 🔁 31 💬 2 📌 1

Inclusive pride flag, showing the rainbow flag with the trans flag and brown and black stripes to represent racial diversity.
I’m sharing my very personal story to encourage everyone to participate in and celebrate Pride month. All our voices are needed to protect diversity and support the queer community. #PRIDEmonth 🌈.
rootandshoot.org/forever-prid...
07.06.2025 17:33 — 👍 53 🔁 16 💬 1 📌 2
Thanks Max!! 😁😁
12.04.2025 19:40 — 👍 1 🔁 0 💬 0 📌 0
A putative 4mC methyltransferase is found in rotifers (N4CMT) that's also thought to have arisen through horizontal gene transfer from bacteria which we talk about briefly in the paper. Wouldn't be surprised if there are others examples out there to be found!
10.04.2025 21:34 — 👍 2 🔁 0 💬 1 📌 0
Thanks Jake!! 😄
10.04.2025 21:31 — 👍 0 🔁 0 💬 0 📌 0
Thanks Sean!! 😁😁😁
10.04.2025 21:30 — 👍 0 🔁 0 💬 0 📌 0
Thanks Li! 😎
10.04.2025 21:30 — 👍 0 🔁 0 💬 0 📌 0
Thanks Tatsuya!! :)
10.04.2025 21:30 — 👍 0 🔁 0 💬 0 📌 0
#DNA #methylation #4mC #epigenetics #germline #Marchantia #sperm #fertility #chromatin #reproduction #plantbiology #evolution
09.04.2025 17:15 — 👍 1 🔁 0 💬 0 📌 0
This work completes a story that started with strange bisulfite-seq anomalies during my PhD and ends with the discovery of a new epigenetic layer in eukaryotic reproduction.
Huge thanks to all co-authors, collaborators, and especially Xiaoqi Feng for guidance throughout.
13/13
09.04.2025 17:15 — 👍 2 🔁 0 💬 1 📌 0
In summary, our study establishes:
4mC is a functional DNA modification in eukaryotes
MpDN4MT1a is a eukaryotic 4mC writer
5mC and 4mC mark distinct chromatin domains in sperm
4mC coordinates transcriptional shutdown, chromatin compaction, sperm motility, and fertility
12/13
09.04.2025 17:15 — 👍 0 🔁 1 💬 1 📌 0
We also propose that 4mC could act as a paternal imprint—for example, guiding PRC2 targeting after fertilization. This may explain why loss of paternal 4mC reduces embryo viability and disrupts development.
11/13
09.04.2025 17:15 — 👍 0 🔁 0 💬 1 📌 0
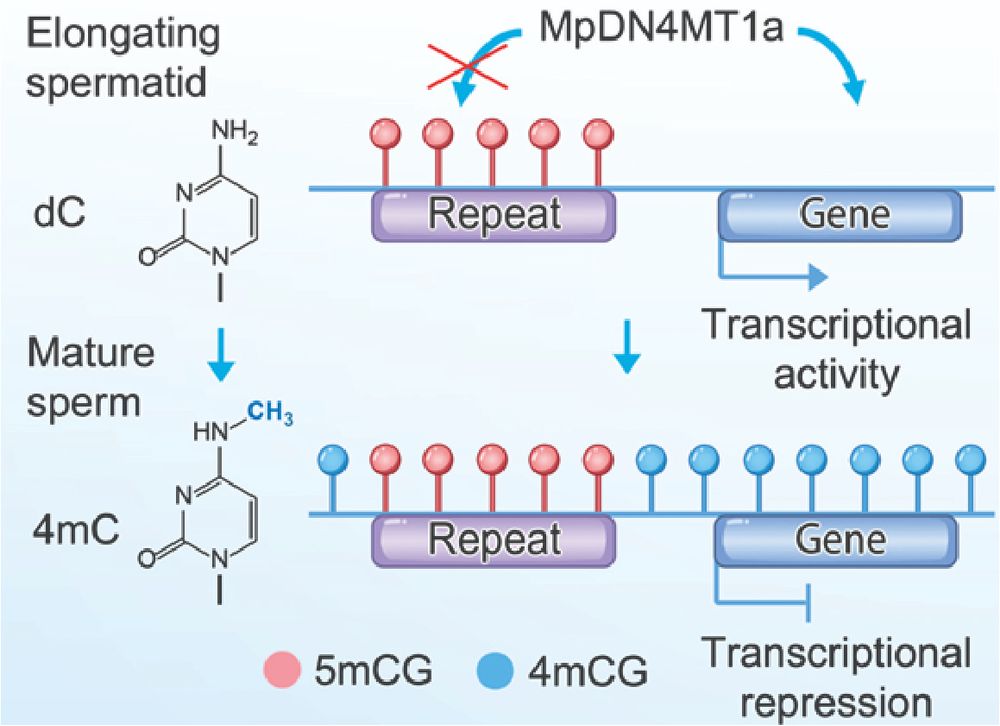
Why deposit 4mC in Marchantia sperm?
We see no evidence of dual-modified 4,5mC, suggesting that 5mC blocks 4mC. This creates a clear division: 4mC marks genes, 5mC marks repeats.
This allows global methylation for compaction while preserving the TE-specific 5mC signature.
10/13
09.04.2025 17:15 — 👍 1 🔁 1 💬 1 📌 0
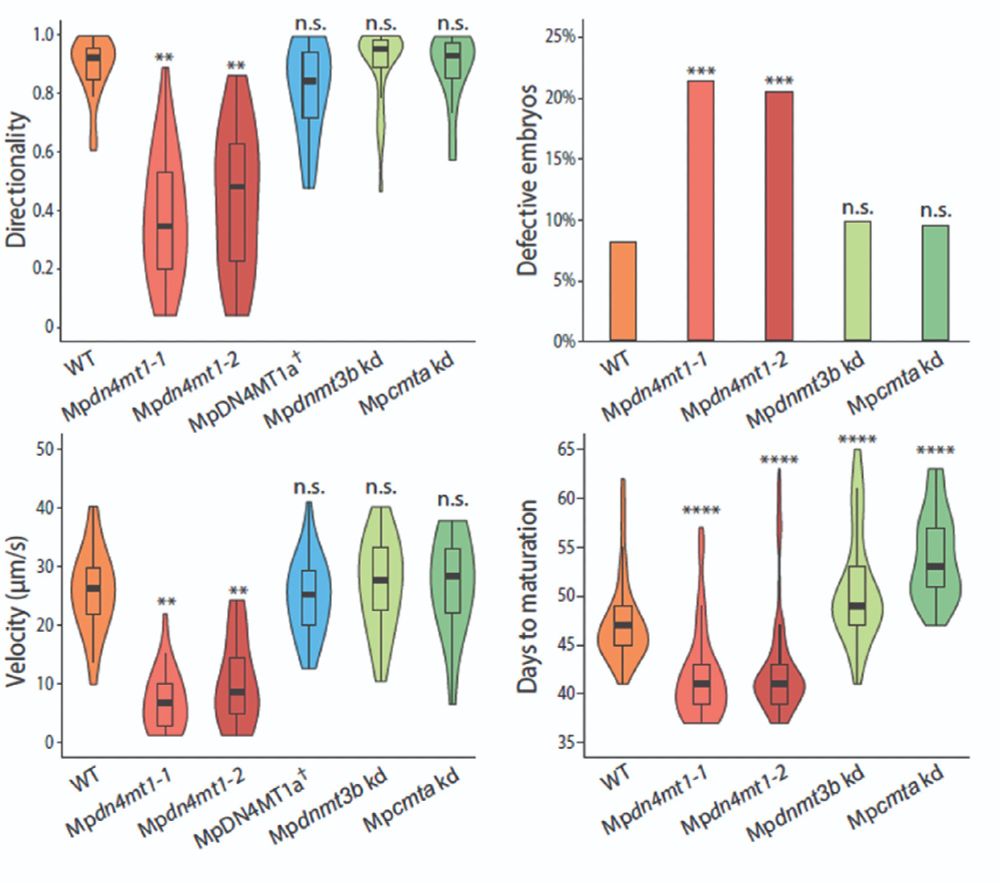
The motility defect is rescued by reintroducing wild-type MpDN4MT1a.
By contrast, sperm from global 5mC mutants show none of these distinctive phenotypes.
9/13
09.04.2025 17:15 — 👍 1 🔁 0 💬 1 📌 0
We now know that 4mC is essential for multiple aspects of sperm function.
Sperm lacking 4mC are motility-defective, outcompeted by wild-type sperm, and produce developmentally compromised embryos.
8/13
09.04.2025 17:15 — 👍 0 🔁 0 💬 1 📌 0
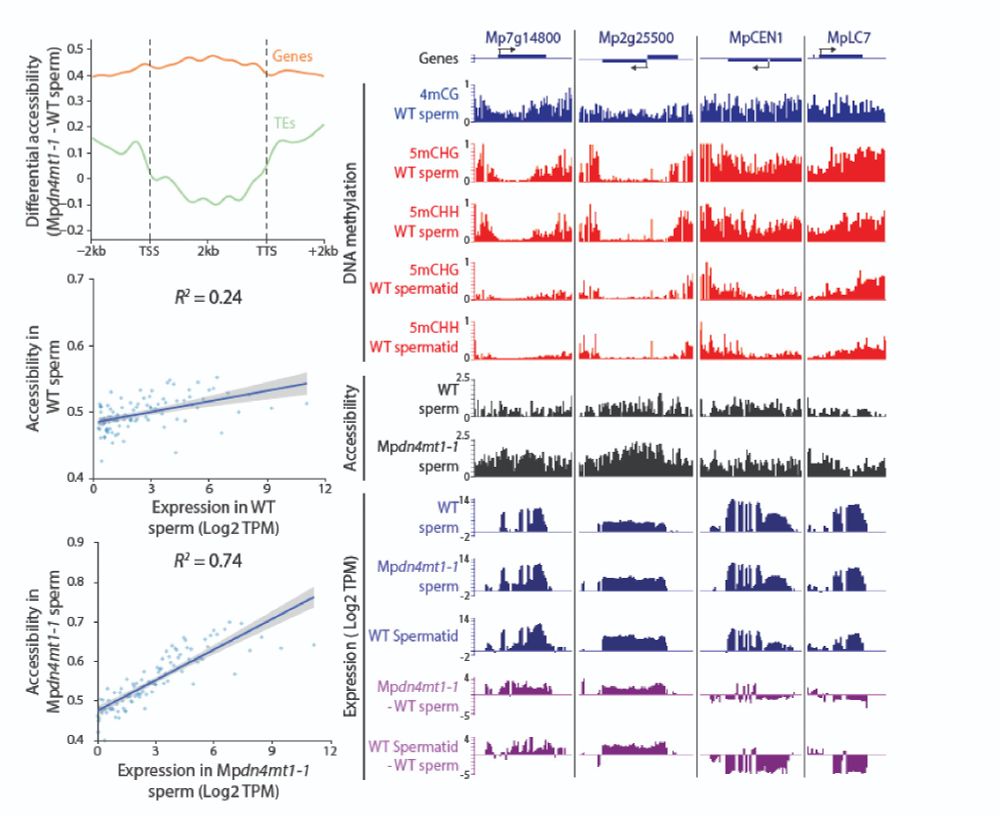
By contrast, transcripts for key sperm function genes—like CENTRIN1 and DYNEIN LIGHT CHAIN 7—are reduced, likely due to a dilution effect, helping explain the sperm motility defect in 4mC mutants.
7/13
09.04.2025 17:15 — 👍 1 🔁 0 💬 1 📌 0
ATAC-seq shows widespread open chromatin in mutants—especially where 5mC is absent at transcription start sites.
Careful RNA-seq analysis uncovered globally elevated expression. As a result, the mutant transcriptome resembles wild-type spermatids before 4mC is established!
6/13
09.04.2025 17:15 — 👍 0 🔁 0 💬 1 📌 0
We previously saw a correlation between 4mC loss and mis-regulated transcription in sperm, but now we uncover the mechanism:
4mC compacts chromatin, silences transcription, and completes the sperm epigenome transition.
5/13
09.04.2025 17:15 — 👍 0 🔁 0 💬 1 📌 0
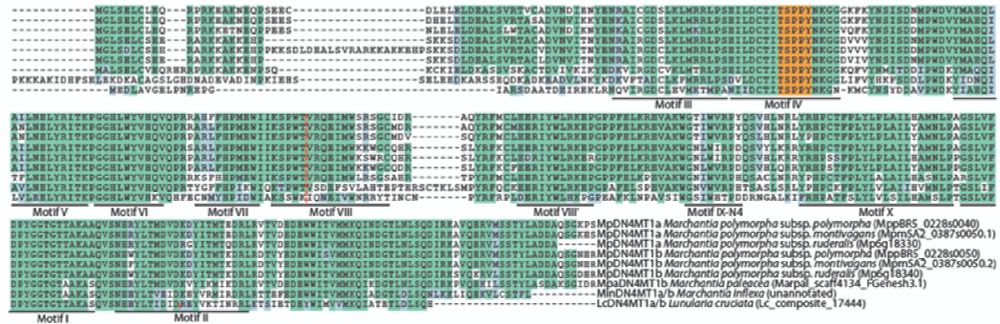
The MpDN4MT1 gene appears to have originated via horizontal gene transfer from prokaryotes and has been retained for at least 200 million years— we detect it even in Lunularia cruciata, a liverwort from a distinct lineage to Marchantia.
4/13
09.04.2025 17:15 — 👍 1 🔁 0 💬 2 📌 0

We show that MpDN4MT1a, a eukaryotic homolog of bacterial 4mC methyltransferases, is the enzyme required for this modification.
Loss of MpDN4MT1a abolishes 4mC, and reintroduction of wild-type—but not the catalytic mutant—restores it.
3/13
09.04.2025 17:15 — 👍 1 🔁 1 💬 1 📌 0
Research group leader @mpi-mp-potsdam.bsky.social
Interested in virus-plant interactions, especially those involving vertical virus transmission from parent to progeny and stem cell antiviral immunity. All posts are my own.
I like science, left politics, concrete architecture and exotic plants. Postdoc at IMBA@Vienna studying Axolotl regeneration and transposons at Elly Tanaka's lab
Biologist studying transposons, gene regulation and chromatin in the Jullien group.
Postdoc @ibmp-cnrs.bsky.social (Strasbourg, France).
Studying MS Biology @LUMS.
Interested in ncRNAs, Genomics, Epigenomics & Gene regulation.
Postdoc in the Brennecke lab @IMBA, Vienna | Alumni: PhD @igbmc @unistra | enthusiast of transcription, chromatin, germline/early development, and transposon biology
Interested in #SmallRNA #Transposons #ParasiticPlants #InterspeciesRNAi #plantscience. Lecturer @sheffielduni.bsky.social. Lab: shahidlab.github.io
Making waves in chromatin @GMIVienna.
Postdoc in the Berger Group at the Gregor Mendel Institute - Vienna BioCenter @gmivienna.bsky.social @viennabiocenter.bsky.social | MSCA Fellow | Interest in evolutionary biology and currently studying chromatin
Plant Biologist 🌱 Junior Professor of Plant Environmental Signalling & Development 🧬🔬 Studying Flooding Tolerance & Epigenetic Memory 🌊 at @biologyunifreiburg.bsky.social @uni-freiburg.de @cibss.bsky.social
www.hartman-plantlab.com
Somatic genomes, DNA repeats, nuclear structure, stem cells and aging - Team leader @i2bcparissaclay.bsky.social - Former @bardinlab.bsky.social
Curious scientist leading a research team mainly working on organelle biology and biotechnology: we study -and often “torture”- chloroplasts to understand how they do their magic ;)
https://www.oeaw.ac.at/gmi/research/research-groups/silvia-ramundo
Group Leader - Reader (Assoc. Professor) in AI and Cancer Epigenetics at Blizard Institute, Queen Mary University of London.
#GeneRegulation, #Chromatin, #Epigenetics, #AI, #bioinformatics
https://www.qmul.ac.uk/blizard/all-staff/profiles/radu-zabet.html
Postdoc in plant epigenetics at University of Oxford, UK in the Mosher lab
Interested in epigenetics 🧬 plant reproduction 🌱 photosynthesis 🌿
Lino print enthusiast 🎨
Plant Physiologist. 🇬🇧in🇺🇸. Loves stomata, plants, and gin
Lecturer in Evolutionary Biology @BristolUni
Evolution of early branching animals
Computational biologist interested in deciphering the genomic regulatory code at vib.ai
Protists, microscopy, biophysics, evolution. Interested in how cells control shape and movement https://www.benlarson.org
Natura in minimis maxima
Asst Prof, RPI | Postdoc, UCSF, Wallace Marshall | PhD, UCBerkeley, Nicole King | BA, Physics, Reed College
Ribozymes, transposable elements, circRNA, viruses, plant biology
Ella-She 💖💜💙♀️🌱
Every fraction of a degree counts
Trans rights are human rights⎥ Black lives matter
Posts in ENG/ESP
🧭 València, Spain
Banner by @jaythechou.bsky.social
Evolutionary cell biology / evolution of morphogenesis / animal origins / choanoflagellates @institutpasteur.bsky.social
https://research.pasteur.fr/en/team/evolutionary-cell-biology-and-evolution-of-morphogenesis/
Professor @uniofgalway.bsky.social. I am interested in developmental biology, stem cells, regeneration, and other things. http://urifranklab.org
chromosome.ie