
For more details about the work on learning uniformly accurate interatomic potentials from scratch I'll present in B1.36:
📄 Paper: www.nature.com/articles/s41...
💻 Code: github.com/nec-research...
@viktorzaverkin.bsky.social
Research Scientist @ NEC Labs Europe, Ph.D. in Theoretical Chemistry @ SimTech & @unistuttgart.bsky.social, ML/DL for Chemistry & Materials Science

For more details about the work on learning uniformly accurate interatomic potentials from scratch I'll present in B1.36:
📄 Paper: www.nature.com/articles/s41...
💻 Code: github.com/nec-research...
Or stop by poster B1.36 (Thu, Aug 28)!
#PsiK2025 #AI4Science
I’ll be at the Psi-k conference next week!
Let’s chat about ML potentials, uncertainty quantification (ensemble-free, gradient-based: Laplace approx., NTKs, batch selection, …), uncertainty-biased MD, message-passing architectures, particle-mesh long-range methods, etc.

🧵 TL;DR:
✅ Benchmark metrics improve with model size and electrostatics
❌ These gains don't always translate to improved simulation outcomes
⚠️ Training data & evaluation practices remain key bottlenecks
📄Preprint: arxiv.org/abs/2508.10841
💻Code: github.com/nec-research...
-/
With @matheusfferraz.bsky.social, @falesiani.bsky.social, @mniepert.bsky.social
15/

Moreover, simulation results are sensitive to training data composition.
E.g., water density predictions depend on whether NaCl-water clusters were in the training set: compare ICTP-LR(M) vs. ICTP-LR(M)*.
Legend: Solid green - ICTP-LR(M); dashed green - ICTP-LR(M)*.
14/
Without DFT-level simulations or other baselines, it is difficult to assess to what extent universal ML potentials improve on classical FFs in realistic biomolecular settings.
While their qualitative advantages are often evident, quantitative validation remains challenging.
13/
These results highlight the limitations of current evaluation practices.
12/

In Trp-cage, simulations with explicit long-range electrostatics exhibit greater conformational variability.
However, the origin of these effects remains unclear without DFT-level simulations.
11/

For Crambin, no significant differences are observed for the vibrational spectrum.
10/

For Ala3, larger models better reproduce experimental J-couplings.
9/

For water and NaCl-water mixtures:
- Larger models don't consistently outperform smaller ones
- Increasing model size doesn't yield systematic convergence
- Explicit electrostatics shifts density predictions from overestimation to underestimation, without consistent gains.
8/
BUT: These improvements do not consistently translate into more accurate physical observables in simulations.
Densities, radial distribution functions, and conformational ensembles show inconsistent trends with model size and long-range electrostatics.
7/

As expected, benchmark metrics (e.g., energy & force RMSEs) systematically improve with increasing model size and the inclusion of explicit long-range interactions.
6/
We use DIMOS for our simulations:
📄Preprint: arxiv.org/abs/2503.20541
💻Code: github.com/nec-research...
5/
We assess the impact of model size, dataset composition, and explicit long-range electrostatics across:
📊 Benchmark datasets
💧 Pure liquid water
🧂 NaCl-water mixtures
🧬 Small peptides (blocked and cationic Ala3)
🧪 Small proteins (Trp-cage, Crambin)
4/
DFT-level simulations and other high-quality baselines are unavailable or infeasible for biomolecular systems.
A more reliable evaluation should consider how model expressivity (model size, explicit long-range interactions) affects prediction errors and simulation results.
3/

📄 Preprint: arxiv.org/abs/2508.10841
💻 Code: github.com/nec-research...
2/

🚨 New preprint: How well do universal ML potentials perform in biomolecular simulations under realistic conditions?
There's growing excitement around ML potentials trained on large datasets.
But do they deliver in simulations of biomolecular systems?
It’s not so clear. 🧵
1/

📎Paper: pubs.acs.org/doi/abs/10.1... (but check also pubs.acs.org/doi/abs/10.1...)
💻Code(s): gitlab.com/zaverkin_v/g... (TensorFlow), github.com/nec-research... (PyTorch), and github.com/apax-hub/apax (JAX)
Many thanks to everyone who has read, cited, or built on it. I hope it continues to be helpful!
25.06.2025 14:42 — 👍 0 🔁 0 💬 1 📌 0We proposed using full tensor contractions to construct many-body features, thereby avoiding expensive sums over triplets, quadruplets, and so on. I am thrilled to see that similar ideas are now an integral part of state-of-the-art architectures, such as MACE, CACE, and so on. 💪
25.06.2025 14:42 — 👍 0 🔁 0 💬 1 📌 0
📈My first PhD paper just reached 100 citations, which is a small but very special milestone for me!
Our paper introduces Gaussian moments as molecular descriptors and uses them to build ML potentials with an impressive balance between accuracy and computational efficiency.
🚀 Apply Now: International Master's Chemical Sciences! 🌍
The application portal for the English-conducted M.Sc. Chemical Sciences at @unistuttgart.bsky.social are officially open! 🔬✨
👉 Visit our program website for further details: www.uni-stuttgart.de/en/study/stu...
I'll present our paper in the afternoon poster session at 4:30pm - 7:30 pm in East Exhibit Hall A-C, poster 3304!
12.12.2024 18:54 — 👍 3 🔁 2 💬 0 📌 0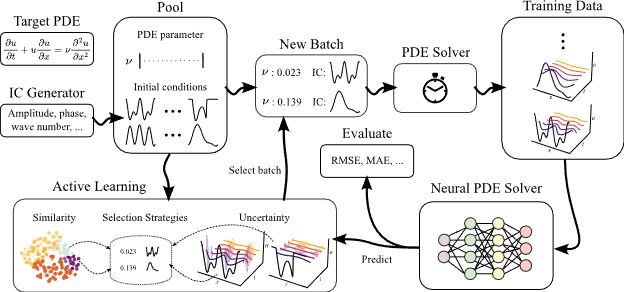
Neural surrogates can accelerate PDE solving but need expensive ground-truth training data. Can we reduce the training data size with active learning (AL)? In our NeurIPS D3S3 poster, we introduce AL4PDE, an extensible AL benchmark for autoregressive neural PDE solvers. 🧵
11.12.2024 18:22 — 👍 12 🔁 3 💬 1 📌 2Join us today at #NeurIPS2024 for our poster presentation:
Higher-Rank Irreducible Cartesian Tensors for Equivariant Message Passing
🗓️ When: Wed, Dec 11, 11 a.m. – 2 p.m. PST
📍 Where: East Exhibit Hall A-C, Poster #4107
#MachineLearning #InteratomicPotentials #Equivariance #GraphNeuralNetworks
@falesiani.bsky.social
11.12.2024 01:20 — 👍 1 🔁 0 💬 0 📌 0@takashimal.bsky.social
08.12.2024 11:03 — 👍 0 🔁 0 💬 1 📌 0Good question! There is a certain connection between Cartesian and geometric products through those operations: geometric product for an n-dimensional vector could be seen as an "outer" product (up to n-rank tensors) + a "contraction". However, I don't know of any systematic study of the relation.
08.12.2024 10:23 — 👍 0 🔁 0 💬 0 📌 0