Most likely only one isoform will be reported, matching the better of the spectra
25.10.2025 19:09 — 👍 0 🔁 0 💬 1 📌 0
Very unlikely to get identified correctly
25.10.2025 17:07 — 👍 0 🔁 0 💬 1 📌 0
Yes, it will. Although without MS/MS evidence the reported localisation confidence will be low and the overall numbers of correctly assigned isomers are likely to be low. Those IDs with MS/MS evidence should be fine though.
25.10.2025 16:42 — 👍 2 🔁 0 💬 1 📌 0

Software Developer
You will contribute to the development of core database applications, web services, data ingestion workflows, and the PRIDE Web platform as part of the PRIDE ecosystem. Working closely with the team, ...
We are #hiring! 🚀
#PRIDE team is looking for a developer to build its future infrastructure. Our focus in a nutshell: #bigdata, single cell proteomics #SCP, & an #AI-oriented platform. Join us to shape the future of open proteomics data!
embl.wd103.myworkdayjobs.com/EMBL/job/Hin...
23.10.2025 13:38 — 👍 10 🔁 5 💬 1 📌 0

The functional landscape of the human ubiquitinome
Protein ubiquitination regulates cell biology through diverse avenues, from quality control-linked protein degradation to signaling functions such as modulating protein-protein interactions and enzyme...
new preprint: Ubiquitin is a protein modification linked with degradation but known to regulate other functions. Over 100k ubiquitination sites have been discovered and here we (@julianvangerwen.bsky.social + others) try to prioritize those most critical to the cell www.biorxiv.org/content/10.1...
09.10.2025 07:07 — 👍 105 🔁 45 💬 2 📌 2
I think this may change with MBR on samples from different patients, in particular for neat.
07.10.2025 17:04 — 👍 1 🔁 0 💬 0 📌 0
That said, it's definitely a good idea for us to provide a tutorial and example code on this. Will add to todo list.
07.10.2025 14:02 — 👍 2 🔁 0 💬 1 📌 0
Currently not saved automatically, main issue: a peptide can match multiple positions even within the sequence of a single protein. However, this can be done with a script, matching to protein sequence in protein_description.tsv output file. AI will easily write such a script in 5 min :)
07.10.2025 14:01 — 👍 2 🔁 0 💬 1 📌 0

Precisely mapping proteases specificity and predicting cleavage rates - with peptide libraries: doi.org/10.1101/2025.... This is a powerful application that is uniquely enabled by the peptidoform confidence and QuantUMS modules in DIA-NN 2.0.
07.10.2025 12:33 — 👍 9 🔁 3 💬 1 📌 0
Do you have an exciting experiment that you'd love to do using proteomics in the context of your work? YPIC funds 5000 eur for your research idea to come to life!
⬇️⬇️⬇️⬇️⬇️
06.10.2025 06:08 — 👍 5 🔁 7 💬 1 📌 0
LinkedIn
This link will take you to a page that’s not on LinkedIn
I will be at the 𝐃𝐈𝐀-𝐍𝐍 session @ 𝐓𝐡𝐞𝐫𝐦𝐨 𝐏𝐫𝐨𝐭𝐞𝐨𝐦𝐞 𝐃𝐢𝐬𝐜𝐨𝐯𝐞𝐫𝐞𝐫 & 𝐂𝐨𝐦𝐩𝐨𝐮𝐧𝐝 𝐃𝐢𝐬𝐜𝐨𝐯𝐞𝐫𝐞𝐫 User Meetings in Bremen, Germany, 𝟗-𝟏𝟏 𝐃𝐞𝐜𝐞𝐦𝐛𝐞𝐫. With some cool advances for DIA-NN on Thermo instruments. www.thermofisher.com/de/de/home/e...
01.10.2025 07:19 — 👍 9 🔁 2 💬 0 📌 0
In DIA-NN 2.3.0, we have added Deamidation (NQ) as an option in the GUI. There are good reasons for it :) You can try InfinDIA with any number of modifications selected, and check the RT differences between modified vs respective stripped peptides, for each mod - a way to validate peptidoform FDR.
29.09.2025 11:46 — 👍 13 🔁 3 💬 0 📌 0
Probably not, we are completely overwhelmed with paper writing at the moment, so only selected stuff eventually will get published. Short application notes are more likely though:)
27.09.2025 08:02 — 👍 1 🔁 0 💬 0 📌 0

With 𝗗𝗜𝗔-𝗡𝗡 𝟮.𝟯.𝟬 Preview (Academia-only for now), we showcase the transformative new capabilities that have been developed in the past months. Download: github.com/vdemichev/Di...
26.09.2025 09:52 — 👍 29 🔁 6 💬 3 📌 0
Start Page: /home/software/Skyline/events/2025 Webinars/Webinar 26
Proteomics Webinar: DIA with FragPipe, DIA-NN, and Skyline
Presenters: Eduard Sabidó and Brendan MacLean
When: Tuesday, September 16, 8am (Pacific Time)
Register Now ... skyline.ms/project/home...
#massspec #proteomics
15.09.2025 08:26 — 👍 33 🔁 16 💬 0 📌 0

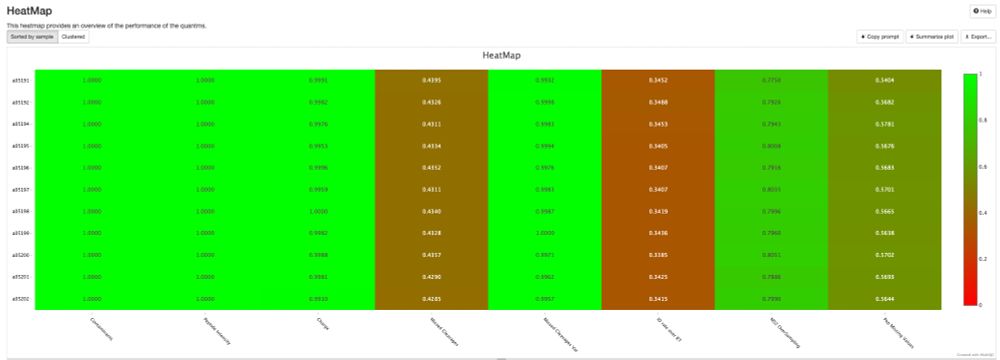
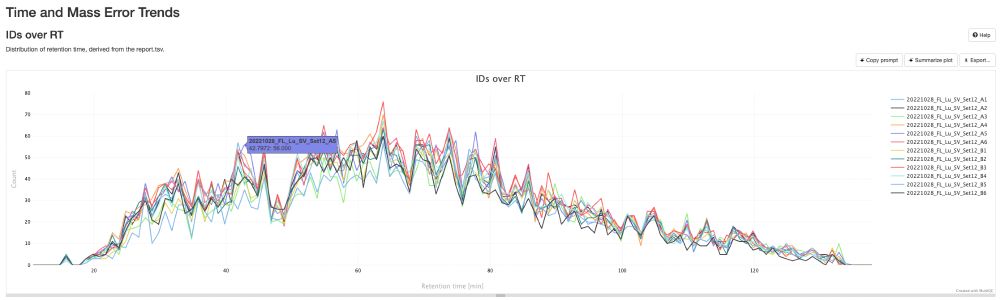
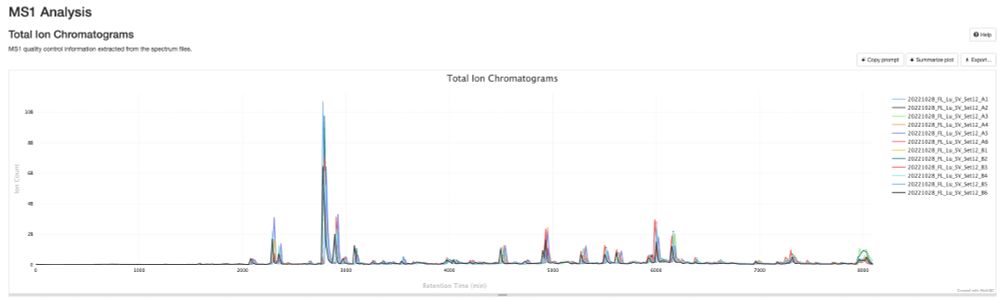
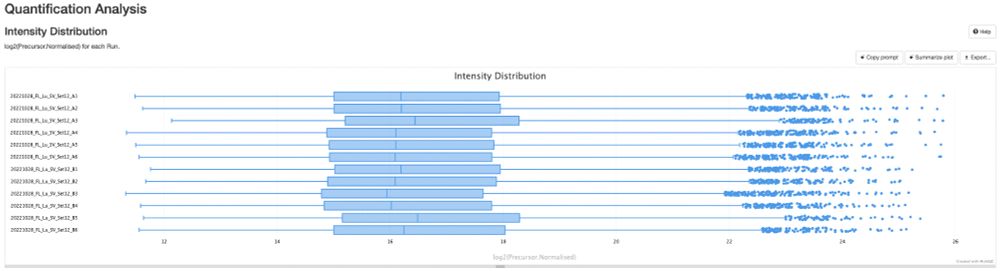
Release v0.0.34 - DIANN and MaxDIA support · bigbio/pmultiqc
What's Changed
fix bugs and add plots for mzid by @yueqixuan in #295
Multiple fix in mzidentml by @ypriverol in #296
MaxQuant: Fix NaN intensity errors by @cbielow in #304
Fix bug in proteobench_u...
New release of pmultiqc v0.0.34 🚀🎉. github.com/bigbio/pmult..., highlights:
- A new contributor, Chris Bielow
- MaxDIA support, thanks @maxquant.bsky.social pmultiqc.quantms.org/MaxDIA/multi...
- DIANN reports way deeper thanks @vadim-demichev.bsky.social pmultiqc.quantms.org/DIANN/multiq...
12.09.2025 12:47 — 👍 7 🔁 4 💬 0 📌 0

Out in @cp-cellsystems.bsky.social: Deep Visual Proteomics shows xenotransplantation drives colon #organoids toward in-vivo-like proteomes supporting their use in regenerative medicine; culture tweaks can mimic this shift. With @kimbakjensen.bsky.social. Lead author Frederik Post explains below:
10.09.2025 15:15 — 👍 19 🔁 3 💬 1 📌 2
Release v2.10.4 · PRIDE-Archive/px-submission-tool
🚀 PX Submission Tool v2.10.4
📦 Downloads
Versioned: px-submission-tool-v2.10.4.zip
Latest: px-submission-tool-latest.zip (always points to current version)
🔗 Universal Links
Latest Release: http...
Major release of @pride-ebi.bsky.social submission tool:
- Smarter Java setup — bundled JRE
- Reliable file transfer logging with Aspera, FTP.
- Clearer instructions and troubleshooting in the README
- Automated releases through GitHub Actions, so updates are consistent
github.com/PRIDE-Archiv...
05.09.2025 09:10 — 👍 3 🔁 3 💬 1 📌 0

We have updated our Slice-PASEF preprint! It's a result of a large collaboration where we highlight its advantages for single cell proteomics. Or any other applications benefiting from sensitivity. For example, major gains for ubiquitinomics:
02.09.2025 17:58 — 👍 19 🔁 3 💬 1 📌 1

Doctoral fellow
PhD Job Alert:
PhD position in metaproteome bioinformatics in Ghent/Belgium with a stay at RKI in Berlin.
www.ugent.be/en/work/scie...
Feel free to share
26.08.2025 21:04 — 👍 2 🔁 1 💬 0 📌 0
On my way to ESCP. Super looking forward to this awesome meeting!
25.08.2025 12:59 — 👍 15 🔁 1 💬 0 📌 0
Postdoc @UoD @CeTPD with Alessio Ciulli
Structural & chemical biology in Targeted Protein Degradation
PhD candidate in statistics at UBC.
Working on statistical genomics.
@biancarsbio's lucky husband.
giulianonetto.netlify.app
Avid Gamer and Coffee Drinker. Not a medical doctor. Co-host to RPG Cave over at Carpool Gaming.
Interested in all things mass spec and glyco 😀
Postdoc & mass spectrometrist. Weekendkost: geen koers = geen friet.
A Taiwanese in Chicago 👨🔬📯🌈🧋🧋🧋
Proteome explorer, horn player and boba enthusiast!
Absea Biotechnology | Empowering protein science
14,000+ recombinant proteins & 11,000+ monoclonal antibodies covering 70% of the human proteome.
Germany-USA-China
https://www.absea.bio/index.html
Shimadzu marks 150 years of innovation in Precision Analytical Instrumentation, focusing on Kaizen & Excellence in Science. Our advanced solutions include Chromatography, Mass Spectrometry & Spectroscopy. Learn more: www.shimadzu.co.uk
Mass spectrometry for high-throughput quantitative proteomics with special interest in posttranslational modifications.
Proteomics at Spanish National Centre for Cardiovascular Research
Chilean scientist exploring mitochondrial proteome homeostasis using (functional) proteomics at the @embl.org. Bridging Excellence Fellow EMBL|Stanford Life Science Alliance.
MSCA doctoral researcher // Erasmus Mundus Scholar // Analytical chemist // Single cell proteomics // Mass spectrometry @ LSMBO, CNRS, Strasbourg, France 🇫🇷
PhD student at @mdc-berlin.bsky.social , studying #metabolism and #geneexpression through #systemsbiology
Fascinated by the regulatory logic behind cell phenotype-environment interactions: How biological cells leverage the structural information encoded in protein molecules to morph themselves into shapes and sizes conferring environmental stress resistance.