🚨 New pre-print! 🚨 In the largest study of its kind to-date, we investigate the ecological and evolutionary mechanisms driving within-patient evolution of antimicrobial resistance (AMR). Read here:
www.biorxiv.org/content/10.6... , and follow along with this thread, discussing our findings (1/21)
20.02.2026 15:57 — 👍 62 🔁 32 💬 2 📌 3

Intracellular competition shapes plasmid population dynamics
From populations of multicellular organisms to selfish genetic elements, conflicts between levels of biological organization are central to evolution. Plasmids are extrachromosomal, self-replicating g...
Hot off the press! Our latest paper led by @fernpizza.bsky.social, understanding how plasmids evolve inside cells. These small, self-replicating DNA circles live inside bacteria and carry antibiotic resistance genes, but also compete with one another to replicate. 1/
www.science.org/doi/10.1126/...
20.11.2025 21:42 — 👍 436 🔁 200 💬 11 📌 18
Our new preprint, led by Martin (@mfenk.bsky.social), is online. We address the microbiome’s role as a source and incubator of acute infections by combining prospective collection of native samples with population-wide, culture-based sequencing. Please check out Martin’s wonderful thread. ⬇️🧵🎉
27.10.2025 11:14 — 👍 11 🔁 3 💬 0 📌 0
This work would have not come alive without many bright heads involved (though many not on BlueSky). Special thanks goes out to my supervisor
@keyfm.bsky.social and our clinical collaborator Bastian Pasieka @torbenbjkd.bsky.social; @iatsenkolab.bsky.social, Carey Nadell, and @mpiib-berlin.mpg.de
27.10.2025 09:24 — 👍 1 🔁 0 💬 1 📌 0
Our results highlight the power of prospective longitudinal investigations of the human microbiome during acute infection leading to yet unknown variants associated with the complex etiology of HAI.
27.10.2025 09:24 — 👍 1 🔁 0 💬 1 📌 0
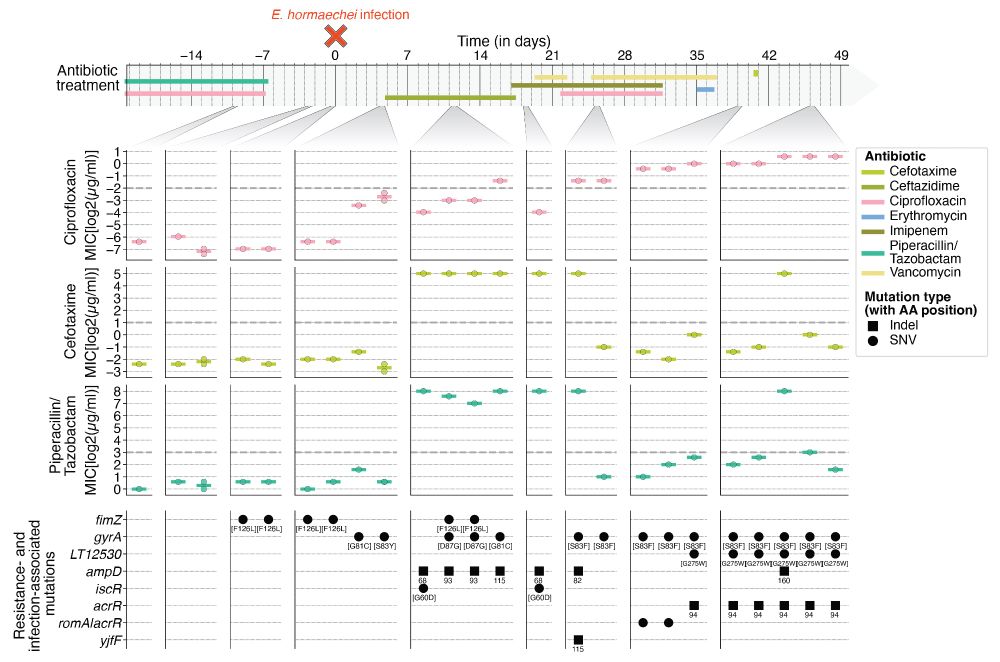
But why was the 𝘧𝘪𝘮𝘡 [F126L] mutant lost shortly after the HAI was cleared? Measuring the antibiotic resistance of isolates over time shows that the replacement was driven by multiple resistance-conferring mutations sweeping across the patient’s body, which were present until the end of the study.
27.10.2025 09:24 — 👍 1 🔁 0 💬 1 📌 0
Intrigued by the infection-associated fitness effects, we wondered whether 𝘧𝘪𝘮𝘡 [F126L] is a common marker of infection. Analyzing >400 global 𝘌. 𝘩𝘰𝘳𝘮𝘢𝘦𝘤𝘩𝘦𝘪 genomes from clinical contexts revealed the 𝘧𝘪𝘮 operon evolves under purifying selection and nonsynonyoums mutations in 𝘧𝘪𝘮𝘡 are rare.
27.10.2025 09:24 — 👍 1 🔁 0 💬 1 📌 0

Further, utilizing 𝘪𝘯 𝘷𝘪𝘷𝘰 survival assays with immunocompromised Relish 𝘋𝘳𝘰𝘴𝘰𝘱𝘩𝘪𝘭𝘢 𝘮𝘦𝘭𝘢𝘯𝘰𝘨𝘢𝘴𝘵𝘦𝘳, we observed an increase in its virulence leading to a more rapid decline in survival rates. This suggests that the 𝘧𝘪𝘮𝘡 [F126L] mutants’ fitness was elevated, potentially beneficial during pneumonia.
27.10.2025 09:24 — 👍 1 🔁 0 💬 1 📌 0

Does the 𝘧𝘪𝘮𝘡 [F126L] mutant convey a fitness effect? To answer this, we performed 𝘪𝘯 𝘷𝘪𝘵𝘳𝘰 assays and found this genotype translates into a hyperpilated phenotype with increased biofilm formation and adherence to lung epithelia.
27.10.2025 09:24 — 👍 2 🔁 0 💬 1 📌 0

During an 𝘌𝘯𝘵𝘦𝘳𝘰𝘣𝘢𝘤𝘵𝘦𝘳 𝘩𝘰𝘳𝘮𝘢𝘦𝘤𝘩𝘦𝘪 infection, we observe a short-lived, non-synonymous mutation in the fimbriae regulator gene 𝘧𝘪𝘮𝘡 [F126L] that is first observed in the gut, before being associated with HAI but subsequently swiftly replaced by repeated body-wide sweeps of independent genotypes.
27.10.2025 09:24 — 👍 3 🔁 1 💬 1 📌 0
The evolutionary trajectory of opportunistic pathogens from asymptomatic carriage to HAI - a critical period for possible pathoadaptation – is rarely studied due to the logistical challenges in prospective sampling. Our population-wide whole-genome sequencing data allowed us to investigate that.
27.10.2025 09:24 — 👍 2 🔁 0 💬 1 📌 0

But were those pathogens acquired within the hospital or outside? Inferring the time to the most common recent ancestor reveals, that some lineages arose from a recent bottleneck during the hospital stay, while others have been colonizing the patient already before hospitalization.
27.10.2025 09:24 — 👍 1 🔁 1 💬 1 📌 0

While we have not seen one common reservoir microbiome niche for those pathogens, we identified that most pathogen strains are observable within the patient’s microbiome either before or within 6 h of HAI onset – pointing at the importance of the microbiome for HAI.
27.10.2025 09:24 — 👍 1 🔁 0 💬 1 📌 0

Of those, 3 patients developed HAIs by 9 different species. We performed culture-based sequencing of each pathogen of ≤10 isolates from the site of infection and ≤20 isolates/microbiome site/timepoint. This uncovered a closely related strain of the pathogens within the microbiome in 73% of cases.
27.10.2025 09:24 — 👍 1 🔁 0 💬 1 📌 0

To gain insights into the hidden evolutionary processes before, during and after HAI, we enrolled 13 critically ill patients and prospectively collected every week native nasal, oral, rectal and skin microbiome samples during their hospital stay.
27.10.2025 09:24 — 👍 2 🔁 0 💬 1 📌 0
Finally getting some of my research for my PhD out there! Hopefully soon in a journal near you ;)
Ancient Y. pestis infection from a sheep falls on the LNBA lineage, previously only known from humans! +molecular evolutionary analysis to leverage the ever increasing number of ancient genomes known!
10.02.2025 17:18 — 👍 15 🔁 10 💬 0 📌 0
Head of the Comparative Functional Genomics lab @Pasteur.fr | @inserm.fr | @erc.europa.eu
Comparative omics - Reproductive evolution - Data witchcraft.
School of Microbiology and APC Microbiome Ireland, University College Cork 🇮🇪
Microbial Ecology | Microbial Evolution | Gut Microbiome
Microbiology, evolution, and bacteriophages. University of Pittsburgh School of Medicine.
PI interested in host-microbe and microbe-microbe interactions in the Drosophila model.
Research group leader at Max Planck Institute for Infection Biology.
https://www.mpiib-berlin.mpg.de/1946812/Genetics-of-Host-Microbe-Interactions
Writing a book about horizontal gene transfer and non treelike evolution. Bioinformatics, Evolutionary Biology. Pangenomes. Chair in Evolutionary Biology. 🇮🇪 http://github.com/mol-evol/panGPT
Wissenschaftliche Vorträge und Stammtisch für Menschen, die an einem rationalen und evidenzbasierten Blick auf die Welt interessiert sind. Am 4. Dienstag des Monats im @weinsalon.bsky.social
Bacterial ecology, evolution and epidemiology | Mathematical and statistical modelling | Genomics | AMR. Assistant professor at the University of Lausanne. https://wp.unil.ch/evolutionaryepidemiology
Group leader at @cnb-csic.bsky.social. Systems biology, complex systems, and spatiotemporal phenomena in life.
All opinions and tweets my own, personal responsibility.
Podcast (Spanish): @lavacaesferica.bsky.social
ORCID: 0000-0001-6214-4083
| CRISPR, Plasmid Biology, Bacterial EcoEvo & AMR | Sci-Illustrator |
Postdoc fellow in Gordo lab @gimminstitute.bsky.social 🇵🇹
Past: @embo.org fellow in San-Millán lab @cnb-csic.bsky.social 🇪🇸 | PhD in Bikard Lab @pasteur.fr 🇫🇷
| She/Her | From 🇪🇸&🇵🇹 | 💜
Evolutionary ecology PhD candidate at Yale University, Patriots fan, and obligately bipedal lobe-finned fish. (he/him)
Ecology and evolution of antibiotic resistance at CNB-CSIC.
@besaraornottobe on X 🐤
Mother, Microbiologist. Crazy about eco-evolutionary aspects of bacteria. #IRYCIS
Microbial ecology and evolution | Professor at University of Lausanne | Theory | Experiments | Egyptian
microbiology | ecology | evolution | biodiversity | lennonlab.github.io
Evo-ecologist (Assoc Prof at UC Berkeley) studying interactions among phages, microbiomes, and plants. Skeets (own view) about science, teaching, social justice, and politics. (profile pic unedited photo by my six-year-old son)
Professor of Microbiome Ecology at Institute of Biology, Leiden University
Bacillus subtilis, biofilms, plant microbiome, bacteria-fungi interactions and circadian clock within @microclockerc.bsky.social #ERCSyG
https://linktr.ee/atkovacs
I play basketball & videogames and do research. Not necessarily in that order. #RyC Group Leader at IPLA-CSIC.
https://msb-lab.github.io/en











