
Thanks to Northwestern for this story behind the story
www.mccormick.northwestern.edu/news/article...
@josh-leonard.bsky.social
My group seeks to grow the field of synthetic biology, with a focus on making advanced gene and cell therapies useful and available to more people. Prof at Northwestern Chem & Biol. Engineering and Center for Synthetic Biology @TheLeonardLab on Twitter

Thanks to Northwestern for this story behind the story
www.mccormick.northwestern.edu/news/article...
Congrats to the whole team, and thanks to NIBIB for leading support of this work! If you're interested in trying these parts, check our page on @addgene.bsky.social or drop me a line!
22.08.2025 18:10 — 👍 0 🔁 0 💬 0 📌 0
Finally we show how these synthetic receptors can be multiplexed to evaluate soluble signals in the environment to create tailored activation programs that can enable "smart" cell therapies
22.08.2025 18:10 — 👍 0 🔁 0 💬 1 📌 0
Examples: cytokine-induced CAR expression on T cells to create a logic gate - the cell is activated only if it sees BOTH a specified soluble cue and surface feature (e.g., tumor antigen), and rewiring cytokine sensing into native pathways that modulate cell state in desired ways
22.08.2025 18:10 — 👍 1 🔁 0 💬 1 📌 0We then leverage the modularity of these converted receptors to link biosensor output to genetic programs, creating new and useful cellular behaviors
22.08.2025 18:10 — 👍 0 🔁 0 💬 1 📌 0
We examine a series of natural receptors, each with a unique (and partially understood) mechanism, and by converting them into synthetic receptors (which we call NatE MESA), we learn the rules governing successful conversions
22.08.2025 18:10 — 👍 0 🔁 0 💬 1 📌 0
How can we combine the exquisite function of natural receptors with the programmability of synthetic biology? Check out this exciting new story led by Hailey Edelstein
and Amparo Cosio, out today in Nature Chemical Biology
nature.com/articles/s41...
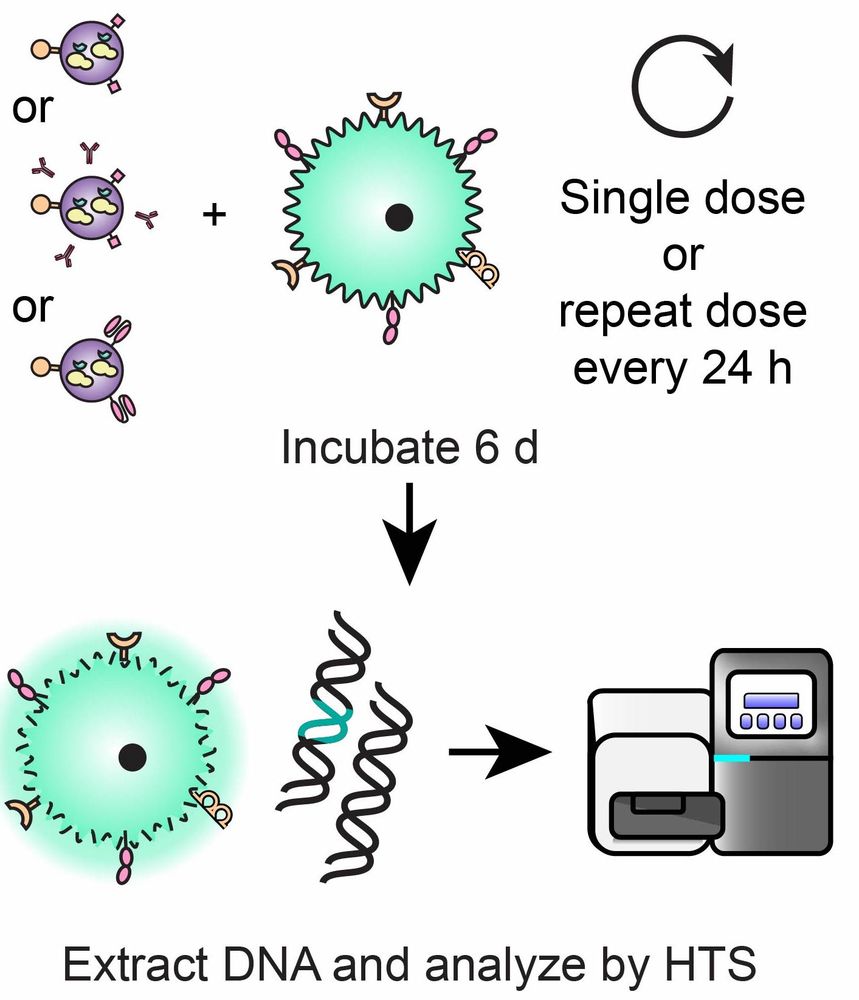
Here we rigorously quantify these challenges (for the first time), develop an assay to overcome this problem, and use this approach to shed new light on delivery phenomena that have previously been elusive and confusing.
More to come with final publication!
However, studying EV-AAV delivery carries unique technical challenges due to co-delivery of AAV and the protein it encodes, termed “pseudotransduction”. This effect can lead to “false” delivery that obfuscates both scientific investigation and technology development.
07.07.2025 13:35 — 👍 0 🔁 0 💬 1 📌 0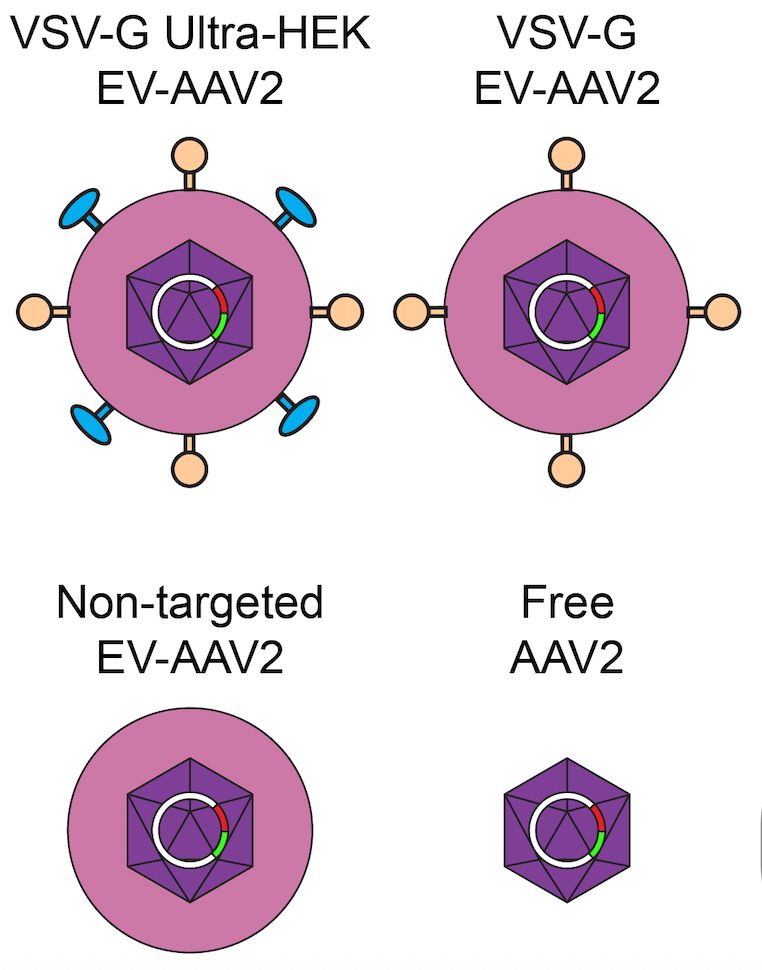
Four types of AAV particle: one is free, the other three are enveloped and are functionalized with affinity domains and fusogens
This can be useful, as enveloping shields the AAV core from antibodies and potentially enables us to use targeting technologies developed for enveloped particles to direct AAV delivery to tissues of interest.
07.07.2025 13:35 — 👍 0 🔁 0 💬 1 📌 0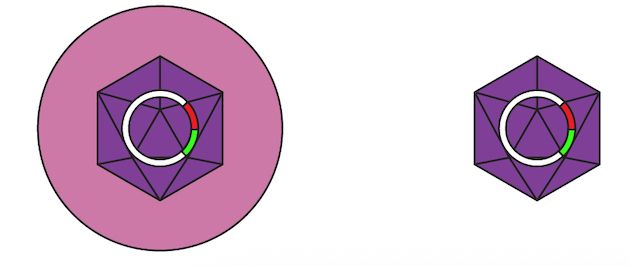
Two AAV particles: one is enveloped and one is free
This preprint describes a new method enabling us to study and improve AAV encapsulated by extracellular vesicles (EVs). When cells produce AAV particles, which normally lack an envelope, a certain fraction get naturally incorporated into EVs.
07.07.2025 13:35 — 👍 0 🔁 0 💬 1 📌 0I'm excited to share this new story led by @jdboucher.bsky.social in which we solve a longstanding problem challenging the development of a promising class of gene delivery vehicle.
www.biorxiv.org/content/10.1...
Deadline to apply was just extended to June 5th!
23.05.2025 17:20 — 👍 0 🔁 0 💬 0 📌 0The original deadline for comments of May 23 has been extended to June 7 in response to requests from the public for more time to weigh in, an OMB spokesperson told STAT.
4/n

Innovate & collaborate at NCI’s S³ Innovation Lab! This virtual event will explore how synthetic & systems biology can advance our understanding of cancer initiation. Apply by May 27: apply.knowinnovation.com/s3-cancer-in... #SynSysBio4SpatialCancerResearch 🧪
23.05.2025 13:32 — 👍 1 🔁 0 💬 1 📌 0
Are you interested in synthetic and systems biology approaches to study #spatiotemporal processes in cancer? Join NCI for virtual workshops on May 13, 15, & 20 (next week!) from 12:00–4:30 PM ET:
registration: events.cancer.gov/nci/syntheti...
#SynSysBio4SpatialCancerResearch

The NSCEB released its final report last week, laying out a bold action plan to shape the future of biotechnology in the US! We look forward to continuing our work with NSCEB and other partners to advance engineering biology and strengthen the bioeconomy! www.biotech.senate.gov/final-report/
14.04.2025 20:03 — 👍 2 🔁 2 💬 0 📌 0 20.12.2024 22:17 — 👍 1 🔁 0 💬 1 📌 0
20.12.2024 22:17 — 👍 1 🔁 0 💬 1 📌 0
👀
17.12.2024 23:25 — 👍 0 🔁 0 💬 0 📌 0
Flyer for a Benzon Symposium on Protein structure prediction and design in biology and pharmacology (Sept. 1–4, 2025). Speakers include: Gabriel Rocklin, Birte Höcker, Amy Keating, Tanja Kortemme, Bruno Correia, Sarel Fleishman, Ashutosh Chilkoti, Minkyung Baek, Noelia Ferruz, James Fraser, Alan Moses, Susan Marqusee, Ben Lehner, Mohammed AlQuraishi, Dek Woolfson, Gustav Oberdorfer, Hannah Wayment-Steele, Ora Schueler-Furman, Jenifer Listgarten, Alexander Rives, & Max Bonomi.
📣 Save the dates 📅
We are organizing a Benzon Symposium on "Protein structure prediction and design" with what I think is an amazing set of speakers
Meeting will take place in Copenhagen 🇩🇰 on Sept. 1–4, 2025, and abstract submission will open in March (benzon-foundation.dk/benzon-sympo...)
Could one envision a synthetic receptor technology that is fully programmable, able to detect diverse extracellular antigens – both soluble and cell-attached – and convert that recognition into a wide range of intracellular responses, from gene expression and real-time fluorescence to modulation..
04.12.2024 16:05 — 👍 434 🔁 139 💬 33 📌 17Exciting to see this beautiful synthetic receptor engineering work out from @piraner #davidbaker (@UWproteindesign) and @KoleRoybal. Congrats all! t.co/t2UuumuTK5
16.11.2024 14:44 — 👍 1 🔁 0 💬 0 📌 0Congrats to lead author DevinStranford and our collaborators Julius Lucks & Judd Hultquist and teams @NUSynBio!
27.11.2023 17:24 — 👍 0 🔁 0 💬 0 📌 0
We believe this technology can be extended to solve all manner of delivery challenges for gene and cell therapy. Check out the story behind the paper and our recent spin-out which is bringing GEMINI to the world through Syenex
news.northwestern.edu/stories/2023...
We demonstrate the utility of GEMINI by delivering Cas9-sgRNA CRISPR complexes to primary human T cells to knock out the gene for CXCR4, one of the receptors that HIV uses to infect cells
27.11.2023 17:23 — 👍 0 🔁 0 💬 1 📌 0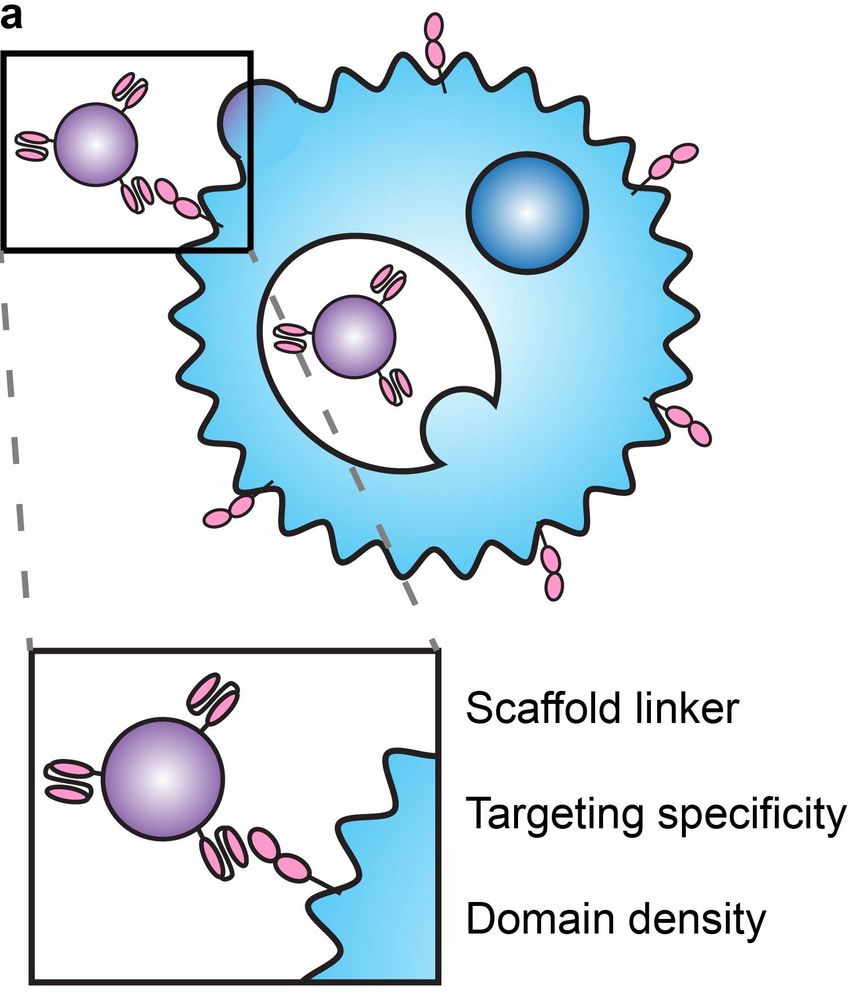
Here we develop a suite of technologies for actively loading cargo into EVs, and we elucidate principles for displaying targeting and fusion domains on the surface of EVs to achieve targeted delivery
27.11.2023 17:22 — 👍 0 🔁 0 💬 1 📌 0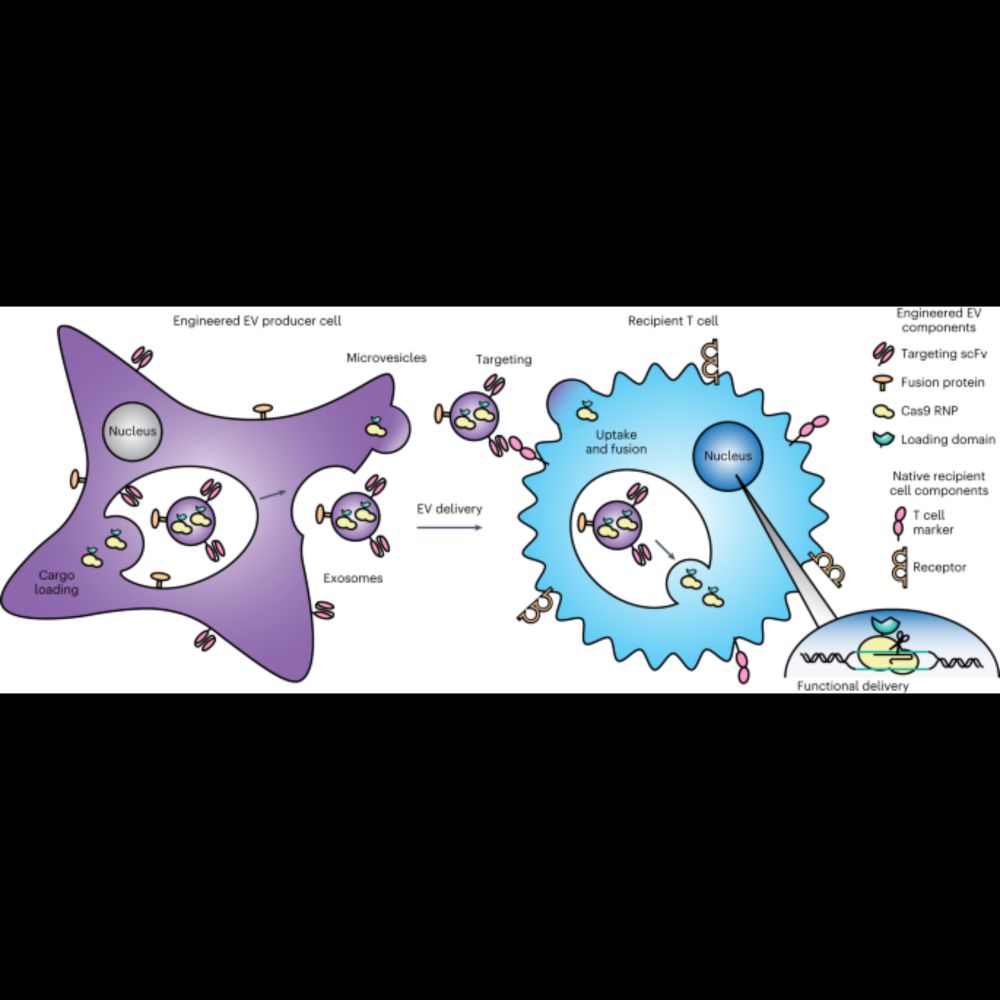
VERY excited to welcome this paper out in @natBME where we introduce the GEMINI platform for engineering multifunctional extracellular vesicles as versatile, programmable, biological delivery vehicles
www.nature.com/articles/s41...
More to come with final publication. Congrats to the whole team! 3/3
27.09.2023 20:23 — 👍 0 🔁 0 💬 0 📌 0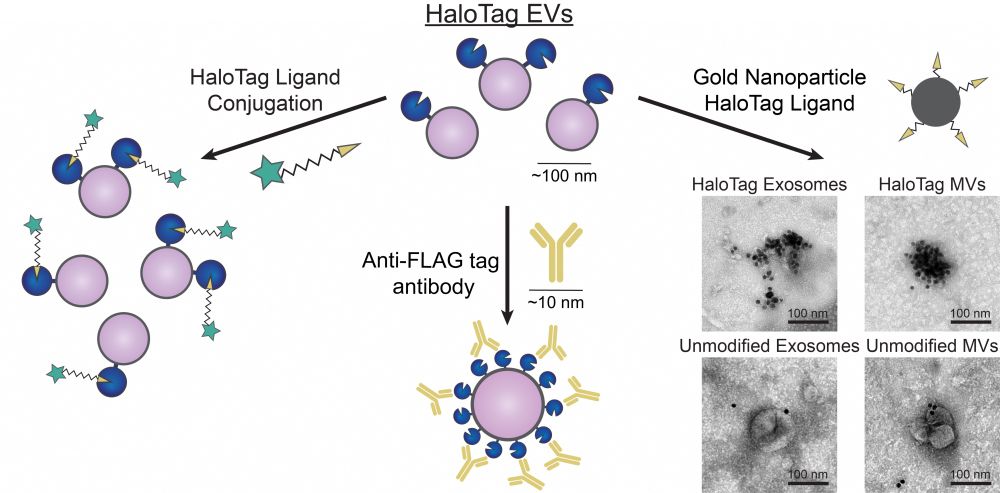
Here we develop a method using flexible HaloTag labeling to absolutely quantify surface protein loading at the individual EV level, providing new insights into EV heterogeneity, and we use this to demonstrate that existing methods substantially underestimate surface display 2/3
27.09.2023 20:13 — 👍 1 🔁 0 💬 1 📌 0
Every cell in the living world constantly sheds virus-size particles, and these “EVs” are useful for biotechnology. Here we address a key unmet need in EV bioengineering research: counting surface features (accurately) 1/3 www.biorxiv.org/content/10.1...
27.09.2023 20:12 — 👍 1 🔁 0 💬 1 📌 0